Abstract
Chemical analysis of the organic extract of Theonella swinhoei yielded two new tridecadepsipeptides of the theonellapeptolide family, namely sulfinyltheonellapeptolide, characterized by a methylsulfinylacetyl group at the N-terminus, and theonellapeptolide If, the first member of this class of compounds to show four valine residues. The structures of the compounds, isolated along with the known theonellapeptolide Id, were determined by extensive 2D NMR and MS/MS analyses followed by application of Marfey’s method. The isolated peptides exhibited moderate antiproliferative activity against HepG2 cells, a hepatic carcinoma cell line.
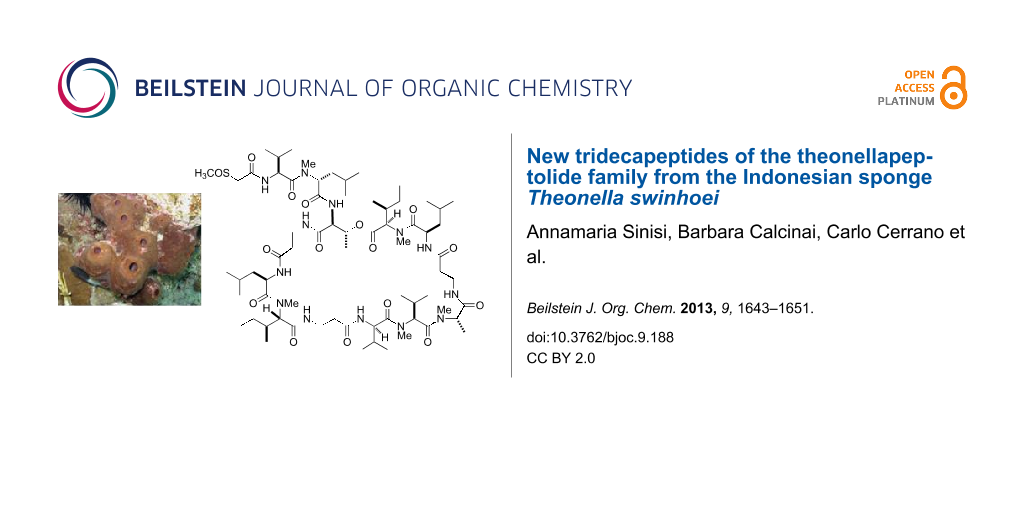
Graphical Abstract
Introduction
Three decades of extensive chemical investigation [1] have clearly evidenced that marine sponges of the genus Theonella (Lithistida, Theonellidae) are treasure troves of secondary metabolites. The chemical diversity of the isolated compounds ranges from unusual steroids (exemplified by the 4-methylene sterol theonellasterol [2,3] and truncated side-chain sulfated steroids [4]), to complex macrocyclic polyketides (as the well-known swinholide A, now a reference compound in the class of actin interacting cell growth inhibitors [5]), polyene derivatives (as aurantosides [6]), and polypeptides/depsipeptides. The biosynthesis of several secondary metabolites of Theonella has been ascribed to symbiotic microorganisms, as in the case of the polyketide onnamide [7] and the polypeptide polytheonamide [8]. It is, however, not unreasonable to presume that a symbiotic role in the production of secondary metabolites could be crucial in many other cases.
Probably the most distinctive class of secondary metabolites of Theonella is given by complex polypeptides whose uncommon amino acids have been postulated to have either nonribosomal (NR) or post-translationally modified ribosomal (PMR) origin [8]. Several classes of Theonella polypeptides have been isolated to date, and they often show peculiar features such as largely rearranged amino acidic units, either D- or L-configurations at the α-carbons, and the formation of macrocycles through amide or ester bonds. To categorize the plethora of Theonella polypeptides we could identify at least eight structural types: theonellamides (glycosylated imidazole-containing macrocycles) [9], keramamides (including oxazole or thiazole rings) [10], papuamides (HIV inhibitory macrocyclic depsipeptides) [11], polytheonamides (cytotoxic linear polypeptides) [12], cyclotheonamides (thrombin and serine protease inhibitors) [13], perthamides (anti-inflammatory cyclopeptides) [14,15], solomonamides [16] and theonellapeptolides (cyclic tridecapeptides including several N-methylated and D-amino acids) [17].
In the context of our long-standing interest for bioactive secondary metabolites from marine invertebrates, we have recently investigated a specimen of T. swinhoei collected off the coasts of Manado (North Sulawesi, Indonesia), which proved to be rich in aurantosides [6] and 4-methylene steroids [18], while polyketide macrolides and peptide-based derivatives were extremely rare if not absent. Remarkably, the chemical analysis of a different specimen of T. swinhoei, collected in the same area as the previous one, exhibited an extremely different secondary metabolite composition. In particular, this second specimen has been found to be rich in polypeptides and, from the CHCl3 phase of the organic extract, we have identified the major member of this class as theonellapeptolide Id (1) (Figure 1). In this paper we report the results of the chemical analysis of the polypeptide-containing fraction, which resulted in the isolation of two new theonellapeptolides, namely sulfinyltheonellapeptolide (2) and theonellapeptolide If (3) (Figure 2). We describe the structural elucidation of these new metabolites and the results of the preliminary pharmacological evaluation of the isolated compounds on HepG2 cells (hepatocarcinoma cell line).
Figure 1: Structure of the known compound theonellapeptolide Id (1).
Figure 1: Structure of the known compound theonellapeptolide Id (1).
Figure 2: Structures of sulfinyltheonellapeptolide (2) and theonellapeptolide If (3).
Figure 2: Structures of sulfinyltheonellapeptolide (2) and theonellapeptolide If (3).
Results and Discussion
Theonella swinhoei was collected in the Bunaken Marine Park of Manado (North Sualwesi, Indonesia) in January 2010 and frozen immediately after collection. The frozen material was repeatedly extracted with methanol, and the crude extract was subjected to a modified Kupchan’s partitioning procedure [19] to obtain n-hexane, CHCl3 and n-BuOH extracts. The CHCl3 extract was chromatographed by silica gel MPLC, and fractions eluted with CH2Cl2/MeOH 97:3 were further purified by HPLC on Nucleodur or Ascentis reversed-phase columns (eluent MeOH/H2O 87:13) to obtain the known theonellapeptolide Id (1, 68.5 mg), and the new analogues sulfinyltheonellapeptolide (2, 4.6 mg) and theonellapeptolide If (3, 2.1 mg) (Figure 2). The known compound was identified on the basis of the comparison of its spectroscopic data with those published in the literature [17].
Theonellapeptolides are tridecapeptide lactones characterized by the presence of aliphatic and non-polar amino acids including high ratio of D-amino acids, N-methyl amino acids, and β-amino acids. In particular, the structure of theonellapeptolide Id (1) includes three β-Ala, three D-Leu (one N-methylated), two L-Val (one N-methylated), one L-MeAla, one L-Thr, one L-MeIle, and two D-alloIle (one N-methylated) units.
Sulfinyltheonellapeptolide (2, [α]D −38.1, c 0.1, MeOH) was isolated as a colorless amorphous solid with pseudomolecular ion peaks at m/z 1423 [M + H]+ and 1445 [M + Na]+ in the ESIMS, and high-resolution analysis established the molecular formula C69H123N13O16S. Inspection of its 1H NMR spectrum (CD3OD, Table 1) clearly suggested the peptide nature of 2 and placed it into the theonellapeptolide class. Given this characterization of 2 the presence of the sulfur atom in the molecular formula appeared especially remarkable. In particular, the 1H and 13C NMR data of 2 indicated the presence of six methyl singlets (δH 2.77, 2.79, 3.22, 3.30, 3.31, and 3.35) and fourteen carbonyl groups (resonating from δC 166.5 to 177.0). Extensive analysis of homonuclear and heteronuclear 2D NMR spectra including 1H/1H COSY, 1H/1H TOCSY, 1H/13C HSQC and 1H/13C HMBC allowed us to overcome the difficulty posed by the severe signal overlap in the 1H NMR spectrum and to establish the presence of three β-Ala, three Leu (one N-methylated), two Ile (both N-methylated), three Val (one N-methylated), one Thr, and one N-methylAla, the same amino acid composition as in theonellapeptolide Id (1) except for one residue. The above analysis left unassigned a methyl singlet at δH 2.79 (δC 39.2) and a pair of mutually coupled doublets at δH 3.67 and 3.86, with these latter signals showing HMBC correlations both with the methyl carbon at δC 39.2 and an amide carbonyl resonating at δC 166.5. The 1H and 13C NMR chemical shift values of both the methyl and the methylene groups strongly suggested their linkage to a sulfoxide group, and the presence of a methylsulfinylacetyl (MeS(O)Ac) subunit was also in full agreement with the molecular formula. The amino acid sequence of 2 was then disclosed by careful analysis of the pattern of HMBC correlations, whose key cross-peaks are shown in Figure 3. The cross-peak of the Thr low-field shifted β-CH (δH 5.19) with the Me-Ile carbonyl group revealed the presence of an ester linkage.
Table 1: 1H (500 MHz) and 13C (125 MHz) NMR data of sulfinyltheonellapeptolide in CD3OD (2).
| AA | Position | δH, m, J in Hz | δC | AA | Position | δH, m, J in Hz | δC |
|---|---|---|---|---|---|---|---|
| L-Thr | 1 | – | 171.2 | β-Ala | 33 | – | 173.2 |
| 2 | 4.36, m | 58.5 | 34/34’ | 2.29, m; 2.59, m | 36.5 | ||
| 3 | 5.19, m | 70.6 | 35/35’ | 3.10, m; 4.23, m | 36.8 | ||
| 4 | 1.11, d, 6.5 | 19.1 | L-Me-Ile | 36 | – | 172.5 | |
| D-Me-Ile | 5 | – | 173.4 | 37 | 3.14, m | 71.2 | |
| 6 | 5.06, m | 62.5 | 38 | 2.48, m | 36.5 | ||
| 7 | 2.14a | 34.2 | 39/39’ | 1.04, m; 1.92a | 30.2 | ||
| 8 | 0.78, d, 7.2 | 11.5 | 40 | 1.00a | 13.2 | ||
| 9/9’ | 1.10, m; 1.38, m | 26.5 | 41 | 0.81, d, 6.4 | 15.3 | ||
| 10 | 0.98, t, 7.6 | 17.0 | 37-NMe | 3.35, s | 39.9 | ||
| 6-NMe | 3.22, s | 32.0 | D-Leu | 42 | – | 176.2 | |
| D-Leu | 11 | – | 177.0 | 43 | 5.06, m | 50.6 | |
| 12 | 5.03, m | 50.5 | 44 | 1.72a; 1.25a | 41.2 | ||
| 13/13’ | 1.74a, 1.25a | 40.4 | 45 | 1.80a | 26.6 | ||
| 14 | 1.81a | 26.3 | 46 | 1.03a | 21.0 | ||
| 15 | 0.89a | 11.1 | 47 | 1.04a | 25.0 | ||
| 16 | 0.94 d, 7.3 | 17.2 | β-Ala | 48 | – | 175.0 | |
| β-Ala | 17 | – | 174.3 | 49/49’ | 2.28, m; 2.36, m | 39.1 | |
| 18/18’ | 2.23, m 2.29 m | 39.5 | 50/50’ | 3.14, m; 3.81, m | 37.7 | ||
| 19/19’ | 3.24, m 3.69, m | 38.2 | D-Me-Leu | 51 | – | 175.2 | |
| L-Me-Ala | 20 | – | 172.5 | 52 | 5.24, m | 57.5 | |
| 21 | 5.17a | 58.5 | 53/53’ | 1.41a; 2.03 m | 39.5 | ||
| 22 | 1.45, d, 7.1 | 16.0 | 54 | 1.52a | 27.1 | ||
| 21-NMe | 2.77 s | 30.5 | 55 | 0.83, d, 6.7 | 22.1 | ||
| L-Me-Val | 23 | – | 173.0 | 56 | 0.95a | 24.3 | |
| 24 | 5.02a | 59.4 | 52-NMe | 3.30 s | 33.0 | ||
| 25 | 2.40, m | 30.2 | L-Val | 57 | – | 176.0 | |
| 26 | 0.86a | 20.6 | 58 | 4.76, m | 57.7 | ||
| 27 | 0.92, d, 6.1 | 20.8 | 59 | 2.12, m | 32.5 | ||
| 24-NMe | 3.31 s | 31.9 | 60 | 0.96a | 20.4 | ||
| D-Val | 28 | – | 173.2 | 61 | 1.01a | 19.2 | |
| 29 | 4.76, m | 68.9 | Me-SAc | 62 | 166.5 | ||
| 30 | 2.68, m | 27.6 | 63/63’ | 3.67, d, 11.2; | 58.9 | ||
| 31 | 0.86a | 18.1 | 3.86, d, 11.2 | ||||
| 32 | 1.17, d, 6.5 | 23.3 | 63-SOMe | 2.79, s | 39.2 | ||
aOverlapped with other signals.
Figure 3: COSY and key HMBC correlations (left) and MS/MS fragmentations of 2 and its ring-opened methanolysis product, respectively.
Figure 3: COSY and key HMBC correlations (left) and MS/MS fragmentations of 2 and its ring-opened methanolysi...
HMBC analysis also revealed that the amide carbonyl at δC 166.5 assigned to the methylsulfinylacetyl subunit was correlated to the Hα of Val-1 residue, thus establishing the acetylation of the N-terminus in 2. Methanolysis of 2, followed by ESI (positive ion mode) MS/MS analysis of the obtained acyclic methyl ester derivative provided key fragment peaks, shown in Figure 3, giving definitive confirmation of the sulfinyltheonellapeptolide gross structure. In addition to the pseudomolecular ion at m/z 1454 [M + H]+, corresponding to the introduction of 32 mass units (MeOH) in the molecule, the ESIMS/MS spectrum provided several fragment ion peaks corresponding to N-terminus fragments due to the cleavage of the amide bond, and referred to as the b series in Roepstorff and Fohlman nomenclature [20]. In particular, the presence of the b2 fragmentation peak at m/z 331 supported, once again, the presence of the MeS(O)Ac unit.
Complete acid hydrolysis of 2 and Marfey’s analysis [21] on the hydrolysate (derivatization with L-FDAA, 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide, followed by LC–MS comparison with the FDAA derivatives of appropriate standards) enabled us to determine the configuration of the chiral amino acid residues as L-MeAla, D-Leu (×2), D-MeLeu, L-Thr, L-Me-Val, L-Me-Ile, D-Me-allo-Ile, D-Val and L-Val. This result left the ambiguity on the localization of the two methylisoleucine residues and of the two enantiomeric valine residues. Since the 1H and 13C NMR resonances for the amino acids at positions 5–10 and 36–41 of 2 were almost superimposable with the corresponding values reported for theonellapeptolide Id (1) [22], we assumed that the two peptides share the configurations at those positions, thus inferring the configurations of the two isoleucine residues. Similarly, it appeared reasonable to assume that, as in the co-occurring theonellapeptolide Id (and in all the theonellapeptolide found to date), the L-Val could be the N-terminal amino acid and thus, the D-Val should be the amino acid at positions 28–32. A D-Val residue at these positions has been found previously in the structure of theonellapeptolide Ia [23]. The configuration at the stereogenic sulfur atom of 2 has been left undetermined.
Theonellapeptolide If (3, [α]D −26.7, c 1.0, MeOH) was isolated as a colorless amorphous solid and its molecular formula was established to be C68H121N13O16 by means of high resolution ESIMS. Inspection of 1H and 13C NMR spectra of 3 (CD3OD, Table 2) revealed extensive similarities with parallel data detected for 2. The most important differences could be recognized in the downfield shift of one methyl singlet (from δH 2.79, δC 39.2 in 2 to δH 3.38, δC 59.1 in 3) and of a pair of mutually coupled doublets (from δH 3.67 and 3.86, δC 58.9 in 2 to δH 3.88 and 3.97, δC 71.8 in 3). These data were easily rationalized with the replacement of the terminal methylsulfinylacetyl group with a methoxyacetyl group, a typical N-terminus acylating unit, already found in the theonellapeptolide family.
Table 2: 1H (500 MHz) and 13C (125 MHz) NMR data of theonellapeptolide If (3) in CD3OD [24].
| AA | Position | δH, m, J in Hz | δC | AA | Position | δH, m, J in Hz | δC |
|---|---|---|---|---|---|---|---|
| L-Thr | 1 | – | 169.2 | β-Ala | 33 | – | 173.2 |
| 2 | 4.38, m | 57.5 | 34/34’ | 2.29, m; 2.59, m | 36.5 | ||
| 3 | 5.15, m | 69.3 | 35/35’ | 3.10, m; 4.17, m | 36.8 | ||
| 4 | 1.13, d, 6.5 | 17.5 | L-Me-Ile | 36 | – | 172.5 | |
| D-Me-Val | 5 | – | 171.5 | 37 | 3.14, m | 71.2 | |
| 6 | 4.95, m | 62.2 | 38 | 2.48, m | 36.5 | ||
| 7 | 2.27a | 26.7 | 39/39’ | 1.04, m; 1.92a | 30.2 | ||
| 8 | 0.76, d, 7.2 | 11.5 | 40 | 1.00a | 13.2 | ||
| 9 | 0.99, d, 7.2 | 23.5 | 41 | 0.81, d, 6.4 | 15.3 | ||
| 6-NMe | 3.17, s | 31.3 | 37-NMe | 3.35, s | 39.9 | ||
| D-Leu | 42 | 176.2 | |||||
| D-Leu | 11 | – | 177.0 | 43 | 5.09, m | 50.6 | |
| 12 | 5.01, m | 48.5 | 44 | 1.72a; 1.25a | 41.2 | ||
| 13/13’ | 1.65a, 1.25a | 40.3 | 45 | 1.78a | 26.6 | ||
| 14 | 1.74a | 24.9 | 46 | 1.03a | 21.0 | ||
| 15 | 0.96a | 11.1 | 47 | 1.04a | 25.0 | ||
| 16 | 0.98 d, 7.3 | 17.2 | β- Ala | 48 | – | 175.0 | |
| β-Ala | 17 | – | 174.3 | 49/49’ | 2.28, m; 2.36, m | 39.1 | |
| 18/18’ | 2.23, m 2.31 m | 39.5 | 50/50’ | 3.14, m; 3.81, m | 37.7 | ||
| 19/19’ | 3.15, m 3.85, m | 38.2 | D-Me-Leu | 51 | – | 175.2 | |
| L-Me-Ala | 20 | – | 172.5 | 52 | 5.18, m | 56.4 | |
| 21 | 5.17a | 58.5 | 53/53’ | 1.41a; 2.03 m | 39.5 | ||
| 22 | 1.45, d, 7.1 | 16.0 | 54 | 1.52a | 27.1 | ||
| 21-NMe | 2.77 s | 30.5 | 55 | 0.83, d, 6.7 | 22.1 | ||
| L-Me-Val | 23 | – | 173.0 | 56 | 0.95a | 24.3 | |
| 24 | 5.02a | 59.4 | 52-NMe | 3.30 s | 33.0 | ||
| 25 | 2.40, m | 30.2 | L-Val | 57 | – | 176.0 | |
| 26 | 0.86a | 20.6 | 58 | 4.98, m | 54.6 | ||
| 27 | 0.92, d, 6.1 | 20.8 | 59 | 2.08, m | 31.6 | ||
| 24-NMe | 3.31 s | 31.9 | 60 | 0.98a | 20.4 | ||
| D-Val | 28 | – | 173.2 | 61 | 1.01a | 19.2 | |
| 29 | 4.76, m | 68.9 | MeO-Ac | 62 | 169.2 | ||
| 30 | 2.68, m | 27.6 | 63/63’ | 3.88, d, 11.2; | 71.8 | ||
| 31 | 0.86a | 18.1 | 3.97, d, 11.2 | ||||
| 32 | 1.17, d, 6.5 | 23.3 | 63-OMe | 3.38 | 59.1 | ||
aOverlapped with other signals.
Careful inspection of COSY and TOCSY spectra revealed also that, in agreement with the molecular formula, one isoleucine residue of 2 is replaced by a valine residue in 3. A subsequent combined analysis of homonuclear and heteronuclear 2D NMR spectra disclosed the presence of the following residues: three β-Ala, three Leu (one N-methylated), one N-methyl-Ile, four Val (two N-methylated), one Thr, and one N-methylAla. As detailed for 2, the amino acid sequence of 3 was deduced by careful analysis of HMBC correlations and supported by the MS/MS spectrum of the methanolysis product mixture (see Supporting Information File 1). Thus, theonellapeptolide If (3) has been identified as a new member of the theonellapeptolide family differing from theonellapeptolide Id (1) by the replacement of two isoleucine residues (positions 5–10 and 28–32) with valine residues. Complete acid hydrolysis of 3 and Marfey’s analysis [21] on the hydrolysate mixture provided the configuration of the chiral amino acid residues as L-MeAla, D-Leu (×2), D-MeLeu, L-Thr, L-Me-Ile, D-Me-Val, D-Val, L-Me-Val and L-Val. With these data in hands, it was not possible to unambiguously define the exact localization of the valine residues. However, if we assume that the L-Val and L-Me-Val residues are localized at the same positions as in theonellapeptolide Id, then the positions of their enantiomeric counterparts are consequently deduced. In summary, compound 3 is a valine-rich theonellapeptolide, being the first member of this class to possess four valine residues.
Theonellapeptolides are structurally similar to the widely used immunosuppressant drug cyclosporin A, sharing at least three important features: i) the exclusive presence of hydrophobic aliphatic amino acids; ii) the abundance of N-methylated residues and iii) the presence of a macrocycle made up by more than ten amino acids. Only a few theonellapeptolides have been tested for immunosuppressive activity and they revealed a moderate potency [25]. In general, the potential bioactivity of this class of unique cyclopeptides has not been investigated in great detail, and an inhibition of the development of fertilized eggs of sea urchin [26], an activity on the transport of Na+ and K+ ions [23], and a moderate cytotoxicity [23] have been reported.
The three theonellapeptolides isolated during the present investigation (1–3) have been evaluated for their antiproliferative activity against HepG2 cells, a hepatic carcinoma cell line. As reported in Figure 4, all tested compounds showed antiproliferative activity at low micromolar doses, with a similar pattern of potency. At the dose 10 μM, the proliferation rate of HepG2 cells was significantly reduced by theonallapeptolide Id (1), sulfinyltheonellapeptolide (2) and theonellapeptolide If (3) to about 50%, 30% and 50%, respectively (Figure 4, panels A, B and C; * p<0.05 compared to untreated cells; n = 4). Only theonellapeptolide Id was able to significantly reduce the proliferation at 1 μM. At this dose, the proliferation reduction was equal to 20% (Figure 4, panel A; * p < 0.05 compared to untreated cells; n = 4). Computation of 50% inhibitory drug concentration (IC50) revealed that compounds 1, 2 and 3 were characterized by very similar IC50 values of about 1.5 μM for theonellapeptolide Id and 3 μM for sulfinyltheonellapeptolide and theonellapeptolide If.
![[1860-5397-9-188-4]](/bjoc/content/figures/1860-5397-9-188-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: Antiproliferative activity of theonellapeptolides 1–3 on hepatic carcinoma cell line. The MTT assay was performed on HepG2 cells treated with increasing doses for 48 hours. Left panel: Proliferation rate expressed as Δ% of absorbance compared to untreated cells. The values are expressed as the mean ± standard error. Right panel: Computation of IC50 values. From top to bottom: Theonellapeptolide Id, sulfinyltheonellapeptolide and theonellapeptolide If (* p < 0.05 compared to untreated cells; n = 4).
Figure 4: Antiproliferative activity of theonellapeptolides 1–3 on hepatic carcinoma cell line. The MTT assay...
Conclusion
In conclusion, the chemical analysis of the organic extract of an Indonesian Theonella swinhoei allowed the isolation of two new members of the theonellapeptolide family, namely sulfinyltheonellapeptolide, characterized by a methylsulfinylacetyl group at the N-terminus, and theonellapeptolide If, the first member of this class to show four valine residues. The structures of the compounds have been determined by extensive 2D NMR and MS/MS analyses followed by the application of Marfey’s method. The position of D- and L-residues was also assigned on the basis of a comparison with known compounds.
A moderate antiproliferative activity on a hepatic carcinoma cell line has been disclosed for the isolated compounds. A more detailed evaluation of the pharmacological profile of this class of marine peptides is underway in our groups.
Experimental
General experimental procedures: Optical rotations were measured on a Perkin–Elmer 243 B polarimeter. Low and high-resolution ESIMS and MS/MS experiments were carried out on a LTQ OrbitrapXL (Thermo Scientific) mass spectrometer. NMR spectra were obtained on Varian Inova 500 MHz spectrometer (1H at 500 MHz, 13C at 125 MHz); δ (ppm), J in Hz, spectra referred to CD3OD (δH 3.32, δC 55.0) as an internal standard. Homonuclear 1H connectivities were determined by COSY experiments. Single-bond heteronuclear 1H/13C connectivities were determined with the HSQC experiment. Two and three bond 1H/13C connectivities were determined by gradient 2D HMBC experiments optimized for a 2,3J = 9 Hz. Medium-pressure liquid chromatography was performed on a Büchi apparatus by using a silica gel (230–400 mesh) column. HPLC was achieved on a Knauer apparatus equipped with a refractive index detector and analytical LUNA (Phenomenex) columns. The purities of compounds were determined to be greater than 95% by HPLC.
Animal material, extraction and isolation: Theonella swinhoei (order Lithistida, family Theonellidae) was collected in the Bunaken Marine Park of Manado (North Sualwesi, Indonesia) in January 2010 and frozen immediately after collection. A reference sample of the sponge has been deposited at the Department of Pharmacy, University of Naples Federico II, with the code Man-10-06. The frozen material (16.5 g) was extracted with methanol (3 × 1.5 L) at room temperature, and the crude methanolic extract was subjected to a modified Kupchan’s partitioning procedure as described in [26]. The CHCl3 extract (4.76 g) was chromatographed with a silica gel MPLC by using a solvent gradient system from CH2Cl2 to CH2Cl2/MeOH 8:2. The fractions eluted with CH2Cl2/MeOH 97:3 (853.3 mg) were further purified by silica gel column chromatography followed by HPLC on a Nucleodur 100-5 C18 (Phenomenex) (5 μm; 7.8 mm i.d. × 250 mm) with MeOH/H2O (9:1) as an eluent (flow rate 0.9 mL/min) to give theonellapeptolide Id (1) (68.5 mg), sulfinyltheonellapeptolide (2, 4.6 mg) and theonellapeptolide If (3, 2.1 mg).
Sulfinyltheonellapeptolide (2): Colorless amorphous solid; [α]D25 −38.0 (c 0.1, CH3OH); 1H and 13C NMR data in CD3OD are given in Table 1; ESIMS: m/z 1423 [M + H]+ and 1445 [M + Na]+. HRMS–ESI (m/z): [M + Na]+ calcd for C69H123N13NaO16S, 1444.8829; found, 1444.8834.
Theonellapeptolide If (3): Colorless amorphous solid; [α]D25 −26.7 (c 0.1, CH3OH); 1H and 13C NMR data in CD3OD are given in Table 2; ESIMS (m/z): 1377 [M + H]+ and 1399 [M + Na]+. HRMS–ESI (m/z): [M + Na]+ calcd for C69H121N13NaO16, 1398.8952; found, 1398.8949.
Acid hydrolysis and Marfey’s analysis: Peptide samples (600 μg) were subjected to acid hydrolysis and subsequent Marfey’s analysis in a similar manner as described in [27]. LC–MS analysis (ESIMS, positive ion mode): individual FDAA-amino acid peak was identified by co-injection with standard amino acid derivatives. The retention times (min) of the FDAA standards were 18.15 for L-Thr, 21.50 for L-MeAla, 29.91 for l-Val, 34.04 for L-MeVal, 34.23 for D-Val, 38.25 for L-MeIle, 34.42 for D-MeVal, 41.35 for D-allo-MeIle, 43.20 for D-Leu, 45.08 for D-MeLeu. Observed amino acids and their retention times (min) for FDAA derivatives of acid hydrolysate of 2 and 3: 2 18.12 for L-Thr, 21.39 for L-MeAla, 29.98 for l-Val, 34.01 for L-MeVal, 34.31 for D-Val, 38.18 for L-MeIle, 41.32 for D-allo-MeIle, 43.19 for D-Leu, 45.01 for D-MeLeu. 3 18.12 for L-Thr, 21.36 for L-MeAla, 29.99 for l-Val, 34.02 for L-MeVal, 34.29 for D-Val, 38.19 for L-MeIle, 34.40 for D-MeVal, 43.17 for D-Leu, 45.02 for D-MeLeu.
Evaluation of antiproliferative activity: HepG2 cells were plated in a 24-wells plate at 3 × 104 cells/well, in Minimum Essential Medium with Earl's salts containing 10% fetal bovine serum (FBS), 1% L-glutamine and 1% penicillin/streptomycin. On day 2, cells were treated with increasing doses of theonellapeptolide Id (0.1, 1 and 10 μM), sulfinyltheonellapeptolide (0.1, 1 and 10 μM) and theonellapeptolide If (0.1, 1 and 10 μM) for 48 hours. On day 4, a MTT assay was assessed: 100 μL of MTT solution (5mg/mL) were added to each well and the cells were incubated at 37 °C for 4 hours. After the incubation, the culture medium was removed and 1 mL of DMSO was added to each well. The absorbance was read by using a spectrophotometer at 590 nM. The proliferation rate was reported as the delta of absorbance compared to untreated cells, and values are expressed as mean ± standard error. Experiments were performed in quadruplicate. For each compound the IC50 value was evaluated as follows: The inhibition of proliferation was expressed as percentage compared to the inhibition of proliferation observed at the highest dose of any experimental setting (which was arbitrarily assigned a value of 100%).
Supporting Information
| Supporting Information File 1: COSY, key HMBC correlations and MS/MS fragmentations of 3, and 1H and COSY spectra for sulfinyltheonellapeptolide (2) and theonellapeptolide If (3). | ||
| Format: PDF | Size: 706.9 KB | Download |
Acknowledgements
This work was supported by the EU project Bluegenics (Grant 311848) and by the University of Naples Federico II (MOU with Universitas Sam Ratulangi, Manado). NMR spectra were recorded at the CSIAS, Centro Interdipartimentale di Analisi Strumentale, Department of Pharmacy, University of Naples Federico II. This research was partially conduced during the Master Course "Tropical Marine Biodiversity and Natural Products" of the Università Politecnica delle Marche.
References
-
Kho, E.; Imagawa, D. K.; Rohmer, M.; Kashman, Y.; Djerassi, C. J. Org. Chem. 1981, 46, 1836–1839. doi:10.1021/jo00322a019
The first paper reporting a chemical investigation on a Theonella sponge dates back to 1981.
Return to citation in text: [1] -
De Marino, S.; Ummarino, R.; D’Auria, M. V.; Chini, M. G.; Bifulco, G.; Renga, B.; D’Amore, C.; Fiorucci, S.; Debitus, C.; Zampella, A. J. Med. Chem. 2011, 54, 3065–3075. doi:10.1021/jm200169t
Return to citation in text: [1] -
Renga, B.; Mencarelli, A.; D'Amore, C.; Cipriani, S.; D’Auria, M. V.; Sepe, V.; Chini, M. G.; Monti, M. C.; Bifulco, G.; Zampella, A.; Fiorucci, S. PLoS One 2012, 7, e30443. doi:10.1371/journal.pone.0030443
Return to citation in text: [1] -
Festa, C.; De Marino, S.; D’Auria, M. V.; Bifulco, G.; Renga, B.; Fiorucci, S.; Petek, S.; Zampella, A. J. Med. Chem. 2011, 54, 401–405. doi:10.1021/jm100968b
Return to citation in text: [1] -
Bubb, M. R.; Spector, I.; Bershadsky, A. D.; Korn, E. D. J. Biol. Chem. 1995, 270, 3463–3466.
Return to citation in text: [1] -
Angawi, R. F.; Bavestrello, G.; Calcinai, B.; Dien, H. A.; Donnarumma, G.; Tufano, M. A.; Paoletti, I.; Grimaldi, E.; Chianese, G.; Fattorusso, E.; Taglialatela-Scafati, O. Mar. Drugs 2011, 9, 2809–2817. doi:10.3390/md9122809
Return to citation in text: [1] [2] -
Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16222–16227. doi:10.1073/pnas.0405976101
Return to citation in text: [1] -
Freeman, M. F.; Gurgui, C.; Helf, M. J.; Morinaka, B. I.; Uria, A. R.; Oldham, N. J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Science 2012, 338, 387–390. doi:10.1126/science.1226121
Return to citation in text: [1] [2] -
Matsunaga, S.; Fusetani, N. J. Org. Chem. 1995, 60, 1177–1181. doi:10.1021/jo00110a020
Return to citation in text: [1] -
Kobayashi, J.; Itagaki, F.; Shigemori, H.; Takao, T.; Shimonishi, Y. Tetrahedron 1995, 51, 2525–2532. doi:10.1016/0040-4020(95)00017-3
Return to citation in text: [1] -
Ford, P. W.; Gustafson, K. R.; McKee, T. C.; Shigematsu, N.; Maurizi, L. K.; Pannell, L. K.; Williams, D. E.; Dilip de Silva, E.; Lassota, P.; Allen, T. M.; Van Soest, R.; Andersen, R. J.; Boyd, M. R. J. Am. Chem. Soc. 1999, 121, 5899–5909. doi:10.1021/JA990582O
Return to citation in text: [1] -
Hamada, T.; Matsunaga, S.; Yano, G.; Fusetani, N. J. Am. Chem. Soc. 2005, 127, 110–118. doi:10.1021/ja045749e
Return to citation in text: [1] -
Fusetani, N.; Matsunaga, S.; Matsumoto, H.; Takebayashi, Y. J. Am. Chem. Soc. 1990, 112, 7053–7054. doi:10.1021/ja00175a045
Return to citation in text: [1] -
Festa, C.; De Marino, S.; D’Auria, M. V.; Monti, M. C.; Bucci, M.; Vellecco, V.; Debitus, C.; Zampella, A. Tetrahedron 2012, 68, 2851–2857. doi:10.1016/j.tet.2012.01.097
Return to citation in text: [1] -
Festa, C.; De Marino, S.; Sepe, V.; D'Auria, M. V.; Bifulco, G.; Andrés, R.; Terencio, M. C.; Payá, M.; Debitus, C.; Zampella, A. Tetrahedron 2011, 67, 7780–7786. doi:10.1016/j.tet.2011.07.077
Return to citation in text: [1] -
Festa, C.; De Marino, S.; Sepe, V.; D'Auria, M. V.; Bifulco, G.; Débitus, C.; Bucci, M.; Vellecco, V.; Zampella, A. Org. Lett. 2011, 13, 1532–1535. doi:10.1021/ol200221n
Return to citation in text: [1] -
Kitagawa, I.; Kobayashi, M.; Lee, N. K.; Shibuya, H.; Kawata, Y.; Sakiyama, F. Chem. Pharm. Bull. 1986, 34, 2664–2667. doi:10.1248/cpb.34.2664
Return to citation in text: [1] [2] -
Angawi, R. F.; Calcinai, B.; Cerrano, C.; Dien, H. A.; Fattorusso, E.; Scala, F.; Taglialatela-Scafati, O. J. Nat. Prod. 2009, 72, 2195–2198. doi:10.1021/np900669d
Return to citation in text: [1] -
Kupchan, S. M.; Britton, R. W.; Ziegler, M. F.; Sigel, C. W. J. Org. Chem. 1973, 38, 178–179. doi:10.1021/jo00941a049
Return to citation in text: [1] -
Roepstorff, P.; Fohlman, J. Biomed. Mass Spectrom. 1984, 11, 601. doi:10.1002/bms.1200111109
Return to citation in text: [1] -
Marfey, P. Carlsberg Res. Commun. 1984, 49, 591–596. doi:10.1007/BF02908688
Return to citation in text: [1] [2] -
Kitagawa, I.; Lee, N. K.; Kobayashi, M.; Shibuya, H. Tetrahedron 1991, 47, 2169–2180. doi:10.1016/S0040-4020(01)96128-9
Return to citation in text: [1] -
Kobayashi, M.; Lee, N. K.; Shibuya, H.; Momose, T.; Kitagawa, I. Chem. Pharm. Bull. 1991, 39, 1177–1184. doi:10.1248/cpb.39.1177
Return to citation in text: [1] [2] [3] -
In the numbering system of theonellapeptolide If (3) we have skipped position 10 to allow for a better comparison between the NMR data of theonellapeptolide If (3) and sulfinyltheonellapeptolide (2).
Return to citation in text: [1] -
Kobayashi, M.; Kanzaki, K.; Katayama, S.; Ohashi, K.; Okada, H.; Ikegami, S.; Kitagawa, I. Chem. Pharm. Bull. 1994, 42, 1410–1415. doi:10.1248/cpb.42.1410
Return to citation in text: [1] -
Gala, F.; D’Auria, M. V.; De Marino, S.; Zollo, F.; Smith, C. D.; Copper, J. E.; Zampella, A. Tetrahedron 2007, 63, 5212–5219. doi:10.1016/j.tet.2007.03.162
Return to citation in text: [1] [2] -
Sepe, V.; D’Orsi, R.; Borbone, N.; D’Auria, M. V.; Bifulco, G.; Monti, M. C.; Catania, A.; Zampella, A. Tetrahedron 2006, 62, 833–840. doi:10.1016/j.tet.2005.10.058
Return to citation in text: [1]
| 26. | Gala, F.; D’Auria, M. V.; De Marino, S.; Zollo, F.; Smith, C. D.; Copper, J. E.; Zampella, A. Tetrahedron 2007, 63, 5212–5219. doi:10.1016/j.tet.2007.03.162 |
| 23. | Kobayashi, M.; Lee, N. K.; Shibuya, H.; Momose, T.; Kitagawa, I. Chem. Pharm. Bull. 1991, 39, 1177–1184. doi:10.1248/cpb.39.1177 |
| 23. | Kobayashi, M.; Lee, N. K.; Shibuya, H.; Momose, T.; Kitagawa, I. Chem. Pharm. Bull. 1991, 39, 1177–1184. doi:10.1248/cpb.39.1177 |
| 1. |
Kho, E.; Imagawa, D. K.; Rohmer, M.; Kashman, Y.; Djerassi, C. J. Org. Chem. 1981, 46, 1836–1839. doi:10.1021/jo00322a019
The first paper reporting a chemical investigation on a Theonella sponge dates back to 1981. |
| 6. | Angawi, R. F.; Bavestrello, G.; Calcinai, B.; Dien, H. A.; Donnarumma, G.; Tufano, M. A.; Paoletti, I.; Grimaldi, E.; Chianese, G.; Fattorusso, E.; Taglialatela-Scafati, O. Mar. Drugs 2011, 9, 2809–2817. doi:10.3390/md9122809 |
| 16. | Festa, C.; De Marino, S.; Sepe, V.; D'Auria, M. V.; Bifulco, G.; Débitus, C.; Bucci, M.; Vellecco, V.; Zampella, A. Org. Lett. 2011, 13, 1532–1535. doi:10.1021/ol200221n |
| 5. | Bubb, M. R.; Spector, I.; Bershadsky, A. D.; Korn, E. D. J. Biol. Chem. 1995, 270, 3463–3466. |
| 17. | Kitagawa, I.; Kobayashi, M.; Lee, N. K.; Shibuya, H.; Kawata, Y.; Sakiyama, F. Chem. Pharm. Bull. 1986, 34, 2664–2667. doi:10.1248/cpb.34.2664 |
| 4. | Festa, C.; De Marino, S.; D’Auria, M. V.; Bifulco, G.; Renga, B.; Fiorucci, S.; Petek, S.; Zampella, A. J. Med. Chem. 2011, 54, 401–405. doi:10.1021/jm100968b |
| 13. | Fusetani, N.; Matsunaga, S.; Matsumoto, H.; Takebayashi, Y. J. Am. Chem. Soc. 1990, 112, 7053–7054. doi:10.1021/ja00175a045 |
| 2. | De Marino, S.; Ummarino, R.; D’Auria, M. V.; Chini, M. G.; Bifulco, G.; Renga, B.; D’Amore, C.; Fiorucci, S.; Debitus, C.; Zampella, A. J. Med. Chem. 2011, 54, 3065–3075. doi:10.1021/jm200169t |
| 3. | Renga, B.; Mencarelli, A.; D'Amore, C.; Cipriani, S.; D’Auria, M. V.; Sepe, V.; Chini, M. G.; Monti, M. C.; Bifulco, G.; Zampella, A.; Fiorucci, S. PLoS One 2012, 7, e30443. doi:10.1371/journal.pone.0030443 |
| 14. | Festa, C.; De Marino, S.; D’Auria, M. V.; Monti, M. C.; Bucci, M.; Vellecco, V.; Debitus, C.; Zampella, A. Tetrahedron 2012, 68, 2851–2857. doi:10.1016/j.tet.2012.01.097 |
| 15. | Festa, C.; De Marino, S.; Sepe, V.; D'Auria, M. V.; Bifulco, G.; Andrés, R.; Terencio, M. C.; Payá, M.; Debitus, C.; Zampella, A. Tetrahedron 2011, 67, 7780–7786. doi:10.1016/j.tet.2011.07.077 |
| 9. | Matsunaga, S.; Fusetani, N. J. Org. Chem. 1995, 60, 1177–1181. doi:10.1021/jo00110a020 |
| 11. | Ford, P. W.; Gustafson, K. R.; McKee, T. C.; Shigematsu, N.; Maurizi, L. K.; Pannell, L. K.; Williams, D. E.; Dilip de Silva, E.; Lassota, P.; Allen, T. M.; Van Soest, R.; Andersen, R. J.; Boyd, M. R. J. Am. Chem. Soc. 1999, 121, 5899–5909. doi:10.1021/JA990582O |
| 8. | Freeman, M. F.; Gurgui, C.; Helf, M. J.; Morinaka, B. I.; Uria, A. R.; Oldham, N. J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Science 2012, 338, 387–390. doi:10.1126/science.1226121 |
| 12. | Hamada, T.; Matsunaga, S.; Yano, G.; Fusetani, N. J. Am. Chem. Soc. 2005, 127, 110–118. doi:10.1021/ja045749e |
| 8. | Freeman, M. F.; Gurgui, C.; Helf, M. J.; Morinaka, B. I.; Uria, A. R.; Oldham, N. J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Science 2012, 338, 387–390. doi:10.1126/science.1226121 |
| 26. | Gala, F.; D’Auria, M. V.; De Marino, S.; Zollo, F.; Smith, C. D.; Copper, J. E.; Zampella, A. Tetrahedron 2007, 63, 5212–5219. doi:10.1016/j.tet.2007.03.162 |
| 7. | Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16222–16227. doi:10.1073/pnas.0405976101 |
| 10. | Kobayashi, J.; Itagaki, F.; Shigemori, H.; Takao, T.; Shimonishi, Y. Tetrahedron 1995, 51, 2525–2532. doi:10.1016/0040-4020(95)00017-3 |
| 27. | Sepe, V.; D’Orsi, R.; Borbone, N.; D’Auria, M. V.; Bifulco, G.; Monti, M. C.; Catania, A.; Zampella, A. Tetrahedron 2006, 62, 833–840. doi:10.1016/j.tet.2005.10.058 |
| 19. | Kupchan, S. M.; Britton, R. W.; Ziegler, M. F.; Sigel, C. W. J. Org. Chem. 1973, 38, 178–179. doi:10.1021/jo00941a049 |
| 6. | Angawi, R. F.; Bavestrello, G.; Calcinai, B.; Dien, H. A.; Donnarumma, G.; Tufano, M. A.; Paoletti, I.; Grimaldi, E.; Chianese, G.; Fattorusso, E.; Taglialatela-Scafati, O. Mar. Drugs 2011, 9, 2809–2817. doi:10.3390/md9122809 |
| 18. | Angawi, R. F.; Calcinai, B.; Cerrano, C.; Dien, H. A.; Fattorusso, E.; Scala, F.; Taglialatela-Scafati, O. J. Nat. Prod. 2009, 72, 2195–2198. doi:10.1021/np900669d |
| 25. | Kobayashi, M.; Kanzaki, K.; Katayama, S.; Ohashi, K.; Okada, H.; Ikegami, S.; Kitagawa, I. Chem. Pharm. Bull. 1994, 42, 1410–1415. doi:10.1248/cpb.42.1410 |
| 23. | Kobayashi, M.; Lee, N. K.; Shibuya, H.; Momose, T.; Kitagawa, I. Chem. Pharm. Bull. 1991, 39, 1177–1184. doi:10.1248/cpb.39.1177 |
| 24. | In the numbering system of theonellapeptolide If (3) we have skipped position 10 to allow for a better comparison between the NMR data of theonellapeptolide If (3) and sulfinyltheonellapeptolide (2). |
| 22. | Kitagawa, I.; Lee, N. K.; Kobayashi, M.; Shibuya, H. Tetrahedron 1991, 47, 2169–2180. doi:10.1016/S0040-4020(01)96128-9 |
| 17. | Kitagawa, I.; Kobayashi, M.; Lee, N. K.; Shibuya, H.; Kawata, Y.; Sakiyama, F. Chem. Pharm. Bull. 1986, 34, 2664–2667. doi:10.1248/cpb.34.2664 |
| 20. | Roepstorff, P.; Fohlman, J. Biomed. Mass Spectrom. 1984, 11, 601. doi:10.1002/bms.1200111109 |
© 2013 Sinisi et al; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (http://www.beilstein-journals.org/bjoc)