Abstract
C-nucleosides have intrigued biologists and medicinal chemists since their discovery in 1950's. In that regard, C-nucleosides and their synthetic analogues have resulted in promising leads in drug design. Concurrently, advances in chemical syntheses have contributed to structural diversity and drug discovery efforts. Convergent and modular approaches to synthesis have garnered much attention in this regard. Among them nucleophilic substitution at C1' has seen wide applications providing flexibility in synthesis, good yields, the ability to maneuver stereochemistry as well as to incorporate structural modifications. In this review, we describe recent reports on the modular synthesis of C-nucleosides with a focus on D-ribonolactone and sugar modifications that have resulted in potent lead molecules.

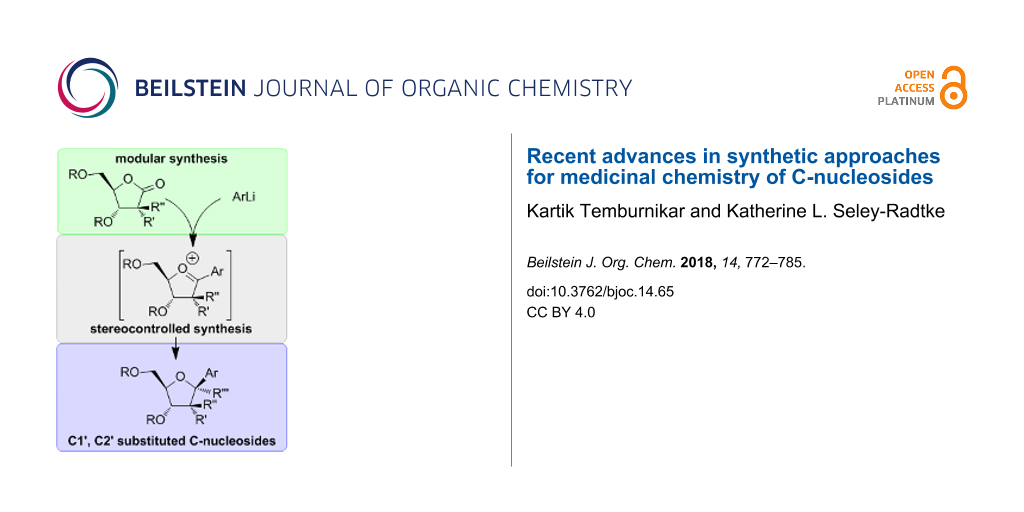
Graphical Abstract
Introduction
Nucleic acids form the genetic blueprint for all living organisms and are involved with a wide range of cellular functions [1-9]. Modifications to their chemical structure can have profound effects on structure and function of enzymes, cells and supramolecular complexes [10-22]. Nucleic acids are composed of a monomeric nucleoside unit that features an aromatic nitrogenous moiety (a nucleobase) connected to a pentose sugar, which in turn is attached to a phosphate group (Figure 1) [7]. The pentose sugar and the nucleobase are connected by a carbon–nitrogen bond that is adjacent to the sugar oxygen resulting in an hemiaminal ether bond, also known as the glycosidic bond.
Figure 1: Structural components of nucleic acids. Shown is the monomeric building block of nucleic acids. Changes to the nucleotide structure can affect molecular recognition, as well as structure and function.
Figure 1: Structural components of nucleic acids. Shown is the monomeric building block of nucleic acids. Cha...
Because of their key role in many biological processes, modifications to the nucleoside structure have been widely employed in the design of drugs, most notably in the fields of virology and cancer research [13-15]. Variations in the nucleoside scaffold are typically accomplished by the insertion, deletion or transposition of functional groups or atoms [23-29]. The varied properties of such modified nucleosides arise from changes in hydrogen bonding motifs, electronic effects, hydrophobic interactions, acid-base properties and chemical reactivity [25-37]. One such modification is the change in the nature of the glycosidic bond [29,37].
Although the glycosidic bond is stable under physiological conditions, cleavage of the bond is common and is highly dependent on the nature of the nucleobase and local pH. In addition, the rate of glycosidic bond cleavage is higher for purines than pyrimdines [38-44]. Moreover, the glycosidic bond in 2'-deoxy ribonucleosides has a higher susceptibility to cleavage than in the corresponding ribonucleosides [38-41,43]. The rate of glycosidic (C–N) bond cleavage is enhanced by decreasing pH and enzymes, which modify the localized acid–base environment [31,35,36]. The C–N bond cleavage proceeds either by activation of a nucleophile that attacks C1' or by stabilization of the leaving group, which could either be the nucleobase or an oxocarbenium ion [31,36]. As such, the oxocarbenium ion is a species formed during the glycosidic bond cleavage, which may be present as an intermediate or a transition state depending upon the accumulation of the positive charge on the sugar ring (Figure 2). As a result, any change in the nucleobase–sugar connectivity (C–N) affects the formation of the oxocarbenium ion and thus influences the stability (or instability) of the nucleoside analogues.
Figure 2: Formation of oxocarbenium ion during glycosidic bond cleavage in nucleosides [31]. The extent of leaving group stabilization and approach of the nucleophile determine charge accumulation on the sugar ring. A concerted process leads to a transition state-like species shown in the figure, while a greater accumulation of positive charge leads to an oxocarbenium ion intermediate.
Figure 2: Formation of oxocarbenium ion during glycosidic bond cleavage in nucleosides [31]. The extent of leavin...
Replacing the hemiaminal (O–C–N) connectivity of the canonical nucleosides with an O–C–C bond (Figure 3) results in a class of compounds called “C-nucleosides” [45-51]. Further modification to a C–C–C connectivity results in “carbocyclic C-nucleosides” (Figure 3) [52,53]. C-nucleosides feature (hetero)aryl aromatic groups such as 9-deazapurines, pyrimidines, pyridines and phenyl groups connected by a C–C bond to a sugar (or sugar mimic) as shown in Figure 4 [30,45-47,50,54-57]. The change in the nature of the glycosidic bond is accompanied by i) increased hydrolytic stability, ii) altered hydrogen bonding motifs, and iii) altered molecular recognition properties [25,29,37,58]. Because of these changes, C-nucleosides have been useful in the study of RNA and DNA processing enzymes, as well as drug design efforts and novel supramolecular structures [12,29,59].
Figure 3: Structural modifications to nucleobase-sugar connectivity. The O–C–N bond between nucleobase and sugar defines the glycosidic bond. Replacement of nucleobase nitrogen by carbon results in C-nucleosides. A carbon (CH2) replacing the sugar oxygen results in carbocyclic nucleosides. When both the heteroatoms in the glycosidic bond are replaced by carbon, the resultant compounds are called carbocyclic C-nucleosides.
Figure 3: Structural modifications to nucleobase-sugar connectivity. The O–C–N bond between nucleobase and su...
Figure 4: Examples of natural and synthetic C-nucleosides. Pseudouridine and formcycin are among several naturally occurring C-nucleosides that are being studied for their role in RNA biology and antibiotic properties respectively. In the recent past, synthetic C-nucleosides, such as immucillin-H and GS-5734, have shown potent activity against purine nucleoside phosphorylases (PNP) and broad spectrum antiviral activities.
Figure 4: Examples of natural and synthetic C-nucleosides. Pseudouridine and formcycin are among several natu...
Pseudouridine is a naturally occurring C-nucleoside that was first discovered in the 1950s [45-47,50]. Subsequently, many more C-nucleosides were discovered and their medicinal properties evaluated (Figure 4) [18,29,37,45-50,60-62]. Due to advances in synthetic methodologies over the years, the repertoire of C-nucleosides has since expanded and has enabled the discovery of clinically useful molecules. Some of the more prominent biologically active analogues that have advanced to clinical evaluations include the immucillins developed by Schramm et al, and Gilead’s antiviral pyrrolo[2,1-f]triazine C-nucleosides (GS-5734 and GS-6620) [32,63-65]. Thus, this review attempts to capture the progress in the synthesis efforts and subsequent drug discovery of the C-nucleosides over the past few years. In the first section, the structural and stereochemical underpinnings of nucleophilic substitutions to D-ribonolactone are discussed, a method that has seen wide applications. Next, we describe reports of different applications and structural variants that have expanded the diversity of the C-nucleosides. Finally, we discuss a modular synthetic approach to carbocyclic C-nucleosides that is also based on the nucleophilic substitution of ribonolactone.
Review
Nucleophilic addition to D-ribonolactone and its stereochemistry
Two prominent methods of C-nucleoside syntheses involve either i) the linear construction of a (hetero)aryl moiety on a C1'-functionalized ribose or ii) coupling of a pre-synthesized (hetero)aryl with a ribosyl moiety (Figure 5A) [48,49,62]. The C–C bond formation usually involves a functional group at C1' of the ribosyl moiety that is amenable to additional functionalization (Figure 5B). Like other nucleoside coupling approaches (other than the well-known Vorbrüggen coupling reaction [66], the synthesis of C-nucleosides typically gives a mixture of stereoisomers (α and β) at the anomeric carbon [48,49,54,62,67,68]. Since the naturally occurring nucleosides (and most biologically active nucleosides) are β-anomers, achieving 100% stereospecificity in C–C bond formation is an important goal, but often difficult to attain [62].
Figure 5: Synthetic approaches to C-nucleosides. A. Two common strategies for C-nucleoside synthesis involve functionalization at C1' and coupling of preformed sugar and heterocylic compounds. B. Structure of ribose and C1' functional groups that enable coupling reactions and synthesis of C-nucleosides.
Figure 5: Synthetic approaches to C-nucleosides. A. Two common strategies for C-nucleoside synthesis involve ...
Among the aforementioned approaches for C-nucleoside syntheses, the coupling of (hetero)aryls to the ribosyl moiety has seen the widest application [52,53,58,62-65,69-77]. This can be ascribed to the modular nature of syntheses that allows for simultaneous alterations in the sugar and the nucleobase to generate diverse analogues in a facile manner. Within this approach, nucleophilic substitution of ribonolactone (Figure 6A) has garnered much attention [61-63,69-75,78]. Ribonolactone typically with its hydroxy groups protected, is amenable to nucleophilic substitutions [78,79]. Use of C-nucleophiles such as lithiated (hetero)aryls leads to C-C bond via a lactol intermediate (Figure 6A). Subsequent deoxygenation of the C1'–OH by Lewis acids (e.g., BF3·OEt2) results in an oxocarbenium ion [62,70,80-83]. Reduction of this intermediate by various silanes gives C-nucleosides resembling the canonical nucleosides [82,83]. The stereochemical fate of oxocarbenium ion reduction is dictated by the conformation and stability of the oxocarbenium ion, which in turn, is affected by the nature of the C2', C3' and C5' substituents [80,81].
Codée and coworkers elaborated on the mechanism and stereochemistry of this reaction by calculating the energies of different oxocarbenium conformers using a free energy surface (FES) mapping method [80,81]. These studies were based on the Woerpel’s model comprising of two stable conformers, namely 3E and E3, in equilibrium (Figure 6B) [84,85]. The nucleophile approaches from the side presenting the least number of eclipsing interactions with the C2' substituent (Figure 6B) [80]. Examining the energies of the various conformers of the permethylated furanosyl oxocarbenium intermediate revealed that the E3 conformer with the C5'–OMe oriented over the positively charged furanosyl ring (Figure 6C) has a large stabilizing effect due to C5'–O5 dipole interactions. In addition, the C2' pseudoequatorial methoxy and C3' pseudoaxial methoxy groups further stabilize the intermediate in E3 conformer, thereby favoring the E3 confomer over the 3E. In the case of an anomeric phenyl group (Ph, Figure 6C), stabilization of the positive charge (C=O+) through conjugation, via parallel alignment, helps to overcome the unfavorable steric interactions between the C2'–OMe and the Ph group [81]. Because E3 is the favored conformer, an inside attack of the nucleophile (H−) results in an α orientation in the final product, which is evident from the the synthesis of 1 (OBn-substituted Pseudouridine, Figure 6D). Despite the greater stability of the E3 conformer, it is the faster reacting conformer (E3 or 3E) that ultimately affects the ratio of diastereomers in the final product [80]. This difference in reactivity results in the differences in various α/β mixtures obtained during the synthesis of C-nucleosides using the D-ribonolactone approach.
Figure 6: Steroselective C-nucleoside synthesis using D-ribonolactone. A. Nucleophilic substitution of D-ribonolactone results in an oxocarbenium ion intermediate. B and C. Functional groups at C2', C3' and C5' stabilize the charged sugar ring and direct the approach of nucleophile to affect the stereochemical outcome of oxocarbenium ion reduction. D. Stereoselective synthesis of protected pseudouridine [80,81].
Figure 6: Steroselective C-nucleoside synthesis using D-ribonolactone. A. Nucleophilic substitution of D-ribo...
Antiviral C-nucleosides
The formation of the lactol and oxocarbenium ion illustrated in Figure 6 also presents the possibility of C1' di-substitution, which was exploited by researchers at Gilead in the discovery of the potent antiviral 4-aza-7,9-dideazaadenine (pyrrolo[2,1-f][1,2,4]triazine) C-nucleosides (Figure 7) [63-65,69,70,77]. The synthesis of 4-aza-7,9-dideazaadenine C-nucleoside 2 (Figure 7A) was first reported by Patil et al. [86] using a sequential approach. In contrast, scientists at Gilead treated perbenzylated ribonolactone with lithiated 4-aza-7,9-dideazaadenine to obtain the lactol intermediate (Figure 7A) [63,65,69,70]. Deoxygenation of the lactol intermediate by BF3·OEt2 resulted in the oxocarbenium ion, which was then reduced using triethylsilane to obtain 2. Replacing triethylsilane with allyl trimethylsilane and trimethylsilyl cyanide gave C1'-allyl (3) and C1'-cyano (4) substitutions respectively. A β/α ratio of 95:5, 87:13 and 89:11 was observed for 2, 3 and 4, respectively, which was sensitive to the reaction temperature and the reagents used [69,70]. A marked difference in diastereomeric purity was observed when the open form of the ribofuranose ring (which exists in equilibrium with the ring closed form), was exploited for C1' substitution using Grignard reagents [69]. Acid-catalyzed dehydration resulted in a diastereomeric mixture of C1'-disubstituted products 5 and 6 with an observed β/α ratio of 2:1 and 1:1, respectively. Similarily, C2'-substituted ribonolactones were employed in the synthesis of 2'-β-Me analogues 7 and 8 and the 2'-deoxy-2'-fluoro 9 (Figure 7B) [65].
Figure 7: Synthesis of C1'-substituted 4-aza-7,9-dideazaadenine C-nucleosides [63-65,69,70]. A. Reaction of D-ribonolactone and lithiated heterocycle leading to C1'-substituted C-nucleosides. Steroechemical scrambling is observed when ring opened form of ribose is employed during nucleophilic substitution. B. Synthesis of C2' C-nucleosides using analogous D-ribonolactones. C. Masked C-nucleoside monophosphates exhibiting potent antiviral activity and clinical utility.
Figure 7: Synthesis of C1'-substituted 4-aza-7,9-dideazaadenine C-nucleosides [63-65,69,70]. A. Reaction of D-ribonolacton...
The 1'-α-H analogue 2 was reported to exhibit inhibitory activity against neoplastic cell lines [86]. This scaffold was later elaborated by Gilead to discover broad spectrum activity of the related C-nucleosides (3–11) against viruses from the Flaviviridae, Orthomyxoviridae, Paramyxoviridae and Coronaviridae families [63-65,69,70]. Cell-based assays revealed potent activity for compounds 2–9 against various viruses including Ebola (EBOV, Filoviridae), respiratory syncytial virus (RSV, Pneumoviridae) and the hepatitis-C virus (HCV, Flaviviridae) family [63,65,69]. Through structure activity relationship studies, the 1'-CN compound 4 emerged as a compound with activity against EBOV, HCV and RSV [65,69]. It is active against EBOV in human microvascular endothelial cells and RSV with low cytotoxicity towards Huh-7, HEp-2 and MT4 cells. Moreover, the triphosphate of 4 selectively inhibits, HCV RdRp and RSV RdRp over human RNA Pol II and DNA polymerases (α, β, γ) [65]. The 2'-Me compound 7 as its triphosphate (TP) shows anti-HCV activity in replicon assays [77]. However, 7-TP serves as a substrate for mitochondrial RNA polymerase, thereby causing toxicity in rats [63]. The 2'-F and 2'-β-Me compounds 8 and 9 are active against the HCV, but lack activity against EBOV and RSV in cell-based assays [65]. The pharmacokinetic properties of 4 were improved by converting it to the masked monophosphates (10 and 11, Figure 7C), which serves to facilitate transport into the infected cells, and conversion to the active triphosphate form, thereby leading to high and persistent levels [63-65]. The 2-ethylbutyl L-alanine phosphoramidate prodrug (Sp isomer, GS-5734, 11) increases the loading of macrophages derived from human monocytes over its unmasked analogue [64]. It was also observed that intravenous administration of the prodrug leads to increased liver loading (as the triphosphate) in hamsters compared to oral dosing [63].
Draffan et al. synthesized a series of 2'-β-Me analogues of pyrrolo- and imidazo[2,1-f][1,2,4]triazine C-nucleosides using a 2'-β-Me lactone that mimic adenosine and guanosine (12–19, Figure 8) [71,72]. The adenine analogues of pyrrolo- and imidazo[2,1-f][1,2,4]triazine were active as nucleosides in HCV1b RNA replication assays, and as triphosphates they inhibit the NS5B polymerase as did the triphosphates of the guanosine analogues [71]. The library of adenosine analogues was further expanded by introducing functional groups at C7 (16–19), which exhibit potent activity in RNA replication assays, with the carboxamide group in particular imparting high potency but also high cytotoxicity [72].
Figure 8: Pyrrolo- and imidazo[2,1-f][1,2,4]triazine C-nucleosides. A series of sugar- and nucleobase-substituted C-nucleosides were synthesized via nucleophilic substitution of D-ribonolactone for structure–activity relationship [71,72].
Figure 8: Pyrrolo- and imidazo[2,1-f][1,2,4]triazine C-nucleosides. A series of sugar- and nucleobase-substit...
A further modification to the imidazo[2,1-f][1,2,4]triazine C-nucleoside scaffold was reported by Dang et al., wherein they synthesized a series of 2'-β-Me analogues possessing a 1',2' cyclopentyl ring (Figure 9) [73]. A representative synthesis (compound 23) is shown in Figure 9, which involves installing an allyl group at C1' (20) and converting the C2'–CN to an aldehyde (21) followed by a Wittig reaction to install a second allyl group at C2' (22). Second generation Grubb’s catalyst was used for the ring formation, followed by hydrogenation to give the desired cyclopentane ring (23) [73]. The biological data of these compounds has yet to be reported.
Figure 9: Synthesis of 1',2'-cyclopentyl C-nucleoside [73]. Functional groups at C1' and C2' were installed and employed for ring cyclization.
Figure 9: Synthesis of 1',2'-cyclopentyl C-nucleoside [73]. Functional groups at C1' and C2' were installed and e...
Wang et al. synthesized a series of pyridine and pyrimidine C-nucleosides (24–26) that mimic the riboside of favipiravir in their effort to develop novel anti-influenza compounds (Figure 10A) [74]. Protected D-ribonolactone 27 was treated with lithiated pyridine to obtain lactol 28 (Figure 10B). Deoxygenation and reduction gave 29, wherein the isopropylidene group was also removed. Conversion of the cyano to an amide group, followed by removal of the silyl protecting group gave 24, which proved to be the most promising compound. The fluorine on 29 was replaced with a methoxy group after re-installing the isopropylidene protecting group. The cyano group was then converted to an amide and the methoxy converted to a hydroxy group. Removal of the protecting groups on the sugar gave 25, which exhibited potent activity against the H1N1 influenza strain (A/WSN/33) in cell based assays [74]. The pyrimidine compound 26 was synthesized using an identical approach and is not shown here. The activity of 24 and 25 as nucleosides was comparable to favipiravir and its riboside. Furthermore, they found that the triphosphate of 24 (24-TP) was incorporated opposite U and C of an RNA template by the influenza polymerase [74]. These experiments indicate that the H-bonding motifs of 24 allow it to mimic both A and G (Figure 10A) [74]. Despite the mis-incorporation, an unmodified sugar moiety may not result in obligate chain termination. While 24-TP is incorporated opposite U and forms more of the full length product than terminated product, its incorporation opposite C results in greater truncated product. Thus, the putative mechanism of action of 24 is through mutagenesis of viral genomic RNA and inhibition of viral polymerase [74].
Figure 10: Anti-influenza C-nucleosides mimicking favipiravir riboside [74]. A. Structure of favipiravir and its riboside, which exhibits anti-influenza activity. C-nucleoside variants of favipiravir riboside and their base pairing with uridine and cytosine. B. Synthesis of C-nucleoside variants of favirpiravir starting from D-ribonolactone.
Figure 10: Anti-influenza C-nucleosides mimicking favipiravir riboside [74]. A. Structure of favipiravir and its r...
Synthesis of C2'-substituted furanolactone
In view of sugar scaffolds possessing C2' substitutions and their value to drug design, a report by Peifer et al. on the synthesis of C2'-substituted ribonolactones is notable (Figure 11A) [75]. Their finding appends known methods of C2' substitution that involve conversion of the C2'-OH to a ketone followed by Me or F substitution [87-94]. Using the Mukaiyama aldol reaction, Peifer obtained a C2'-substituted ribonolactone, which can then be employed in C-nucleoside synthesis [75]. This involves condensation of alkyl-substituted silyl ketene acetals (32) with enantioenriched α-2,2,6,6-tetramethylpiperidinyl-β-benzyloxypropionaldehyde (33) in presence of TiCl2(OiPr)2 to give the β-hydroxyester 34 that is diastereomerically enriched [75,95]. Reductive cleavage of the 2,2,6,6-tetramethylpiperidinyl (TMP) group by Zn and trifloroacetic acid results in cyclization and formation of the C2'- substituted ribonolactone (35). TiCl2(OiPr)2 has been identified as the optimal Lewis acid for the synthesis of most ribonolactones with the exception of unsubstituted silyl ketene acetals (R = R' = H) that leads to stereochemical inversion at C3' [75]. The desired stereoselectivity for 2'-deoxy analogues was obtained when BF3·OEt2 was used. Furthermore, another route to the synthesis of C-nucleosides was demonstrated by direct addition of aryl lithium reagents to the 2'-OMe ribonolactone (Figure 11B). While the expected lactol was formed, deoxygenation by BF3·OEt2 and reduction in presence of the Hantzsch ester afforded the desired β-anomer, while the use of Et3SiH gave the α-anomer [75].
Figure 11: Alternative method for synthesis of 2'-substituted C-nucleosides [75]. A. Synthesis of C2'-substituted D-ribonolactone via Mukaiyama aldol reaction. A series of 2'-O-alkyl, alkyl, cycloalkyl and deoxy D-ribonolactone were synthesized using this method. B. Use of Hantzsch ester to obtain the β-anomer of C-nucleosides.
Figure 11: Alternative method for synthesis of 2'-substituted C-nucleosides [75]. A. Synthesis of C2'-substituted ...
Carbocyclic C-nucleosides
In an attempt to synthesize carbocyclic C-nucleosides, Maier et al. found that reaction of aryl lithiums with pentanone 37 results in carbocyclic C-nucleosides with a C1'-hydroxy group (38 and 39, respectively, Figure 12A) [52,53]. They synthesized cyclopentanone 37 in 7 steps starting from norbornadiene (40, Figure 12B). Furthermore, silyl protection (TIPS) of the C2' and C3' was observed to be critical for the stability of 37 and to facilitate functional group interconversions as shown in Figure 12A [53].
Figure 12: Synthesis of carbocyclic C-nucleosides using cyclopentanone [53]. A. Nucleophlic substitution on cyclopentanone gives C1'–OH carbocyclic C-nucleosides. B. Synthesis of cyclopentanone from norbornadiene and substituents that facilitate carbocyclic C-nucleoside syntheses.
Figure 12: Synthesis of carbocyclic C-nucleosides using cyclopentanone [53]. A. Nucleophlic substitution on cyclop...
In order to obtain carbocyclic C-nucleosides that resemble canonical nucleosides, Maier and coworkers synthesized a stable enol triflate (46, Figure 13A), which then enables Suzuki coupling and a modular synthesis of carbocyclic C-nucleosides [53]. The boronic acids/boronates (inset, Figure 13) of several (hetero)aryls were conducive to Suzuki coupling with the best result obtained when the C2', C3' and C5'–OHs were protected with TIPS and pivaloyl groups, respectively [53]. The cross-coupling reaction gave the unsaturated compounds (47 and 48), which, upon hydrogenation in presence of Crabtree’s catalyst, gave the saturated compounds with the desired diasteroselectivity (49 and 50). In the case of nitrogen containing heterocycles, Pd(OH)2 was found to be a suitable catalyst that gave a separable mixture of diastereomers (2:1). In addition, optically pure cyclopentanone (−)-45 was obtained by converting the cyclopentane 43 to camphanates (51, Figure 13B) followed by separation of the diastereomers [53]. Subsequent synthesis of the enol triflate (−)-46 (Figure 13B), Suzuki coupling and hydrogenation afforded the optically pure carbocyclic tubercidine analogue (−)-53. This compound has shown potent activity against breast cancer cell lines and human foreskin fibroblasts [53].
Figure 13: Synthesis of carbocyclic C-nucleosides via Suzuki coupling [53]. A. Synthesis of OTf-cyclopentene that enable Suzuki coupling and modular synthesis of carbocyclic C-nucleosides. B. Synthesis of enantiomerically pure 4-aza-7,9-dideazaadenine carbocyclic C-nucleoside.
Figure 13: Synthesis of carbocyclic C-nucleosides via Suzuki coupling [53]. A. Synthesis of OTf-cyclopentene that ...
Conclusion
With increasing reports of emerging and reemerging infectious diseases globally, there is a need to develop more effective and safer drugs. In that regard, C-nucleosides have recently shown great potential, which in turn, has resurrected interest in this class of molecules [29]. Several antiviral C-nucleosides have been discovered in the past five years and are now in advanced stages of clinical applications. The overarching features of these compounds with regards to changes in the nucleobase and sugars allow optimal interactions with enzymes resulting in potent and often times, selective, inhibitory activities [18,65,74,96]. As continuing efforts to design greater diversity in C-nucleosides, methods of their synthesis have become critical to more effective drug discovery. For example, the pyrrolo[2,1-f][1,2,4]triazine scaffold has been key to the discovery of several highly active molecules [53,69,71-73,86]. Modular and convergent synthetic routes have proved valuable in this regard both in terms of increasing diversity and reducing the time and length of the syntheses [70-73,76]. Efforts have been aided by advances in the synthesis of modified sugars and sugar mimics, particularly D-ribonolactone analogues [53,73,75,97]. Furthermore, chemical and theoretical studies have elucidated the mechanism and stereochemical preferences of reactions involving D-ribonolactone [80,81,84,85]. Therefore, the chemist has better control over the reactions with more predictable outcomes. In the coming years, new applications may be reported. Moreover, with the biological potential of C-nucleosides now being revisited, studies of naturally occurring C-nucleosides and their biosynthetic pathways have garnered renewed interest, as has the pursuit of new biosynthetic C-nucleosides [98-104]. Previously reported C-nucleosides are also being revisited and may be repurposed with increased knowledge of new biological targets [29,65,86,96]. In summary, these efforts, in concert with improved synthetic advances, provide strong impetus for the next wave of C-nucleoside design and the discovery of nucleoside therapeutics.
References
-
Miescher, F. Med. Chem. Unters. 1871, 441–460.
Return to citation in text: [1] -
Avery, O. T.; Macleod, C. M.; McCarty, M. J. Exp. Med. 1944, 79, 137–158. doi:10.1084/jem.79.2.137
Return to citation in text: [1] -
Chargaff, E.; Lipshitz, R. J. Am. Chem. Soc. 1953, 75, 3658–3661. doi:10.1021/ja01111a016
Return to citation in text: [1] -
Franklin, R. E.; Gosling, R. G. Nature 1953, 172, 156–157. doi:10.1038/172156a0
Return to citation in text: [1] -
Watson, J. D.; Crick, F. H. C. Nature 1953, 171, 737–738. doi:10.1038/171737a0
Return to citation in text: [1] -
Watson, J. D.; Crick, F. H. C. Nature 1953, 171, 964–967. doi:10.1038/171964b0
Return to citation in text: [1] -
Nelson, D. L.; Cox, M. M. Lehninger Principles of Biochemistry, 4th ed.; Freemand and Company, 2005.
Return to citation in text: [1] [2] -
Matera, A. G.; Terns, R. M.; Terns, M. P. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. doi:10.1038/nrm2124
Return to citation in text: [1] -
Rhodes, D.; Lipps, H. J. Nucleic Acids Res. 2015, 43, 8627–8637. doi:10.1093/nar/gkv862
Return to citation in text: [1] -
Hamma, T.; Ferre-D'Amare, A. R. Chem. Biol. 2006, 13, 1125–1135. doi:10.1016/j.chembiol.2006.09.009
Return to citation in text: [1] -
McDonald, M. K.; Miracco, E. J.; Chen, J.; Xie, Y.; Mueller, E. G. Biochemistry 2011, 50, 426–436. doi:10.1021/bi101737z
Return to citation in text: [1] -
Boettcher, T.; Sieber, S. A. J. Am. Chem. Soc. 2010, 132, 6964–6972. doi:10.1021/ja909150y
Return to citation in text: [1] [2] -
Jordheim, L. P.; Durantel, D.; Zoulim, F.; Dumontet, C. Nat. Rev. Drug Discovery 2013, 12, 447–464. doi:10.1038/nrd4010
Return to citation in text: [1] [2] -
Coats, S. J.; Garnier-Amblard, E. C.; Amblard, F.; Ehteshami, M.; Amiralaei, S.; Zhang, H.; Zhou, L.; Boucle, S. R. L.; Lu, X.; Bondada, L.; Shelton, J. R.; Li, H.; Liu, P.; Li, C.; Cho, J. H.; Chavre, S. N.; Zhou, S.; Mathew, J.; Schinazi, R. F. Antiviral Res. 2014, 102, 119–147. doi:10.1016/j.antiviral.2013.11.008
Return to citation in text: [1] [2] -
Pradere, U.; Garnier-Amblard, E. C.; Coats, S. J.; Amblard, F.; Schinazi, R. F. Chem. Rev. 2014, 114, 9154–9218. doi:10.1021/cr5002035
Return to citation in text: [1] [2] -
Wiegmann, D.; Koppermann, S.; Wirth, M.; Niro, G.; Leyerer, K.; Ducho, C. Beilstein J. Org. Chem. 2016, 12, 769–795. doi:10.3762/bjoc.12.77
Return to citation in text: [1] -
Serpi, M.; Ferrari, V.; Pertusati, F. J. Med. Chem. 2016, 59, 10343–10382. doi:10.1021/acs.jmedchem.6b00325
Return to citation in text: [1] -
Maffioli, S. I.; Zhang, Y.; Degen, D.; Carzaniga, T.; Del Gatto, G.; Serina, S.; Monciardini, P.; Mazzetti, C.; Guglierame, P.; Candiani, G.; Chiriac, A. I.; Facchetti, G.; Kaltofen, P.; Sahl, H.-G.; Dehò, G.; Donadio, S.; Ebright, R. H. Cell 2017, 169, 1240–1248. doi:10.1016/j.cell.2017.05.042
Return to citation in text: [1] [2] [3] -
Davis, J. T.; Spada, G. P. Chem. Soc. Rev. 2007, 36, 296–313. doi:10.1039/B600282J
Return to citation in text: [1] -
Jones, M. R.; Seeman, N. C.; Mirkin, C. A. Science 2015, 347, 1260901. doi:10.1126/science.1260901
Return to citation in text: [1] -
Krishnan, Y.; Simmel, F. C. Angew. Chem., Int. Ed. 2011, 50, 3124–3156. doi:10.1002/anie.200907223
Return to citation in text: [1] -
Fenniri, H.; Temburnikar, K. W.; Johnson, R. S. In Comprehensive Supramolecular Chemistry II; Atwood, J. L., Ed.; Elsevier: Oxford, 2017; pp 83–113. doi:10.1016/B978-0-12-409547-2.12583-1
Return to citation in text: [1] -
Cihak, A. Oncology 1974, 30, 405–422. doi:10.1159/000224981
Return to citation in text: [1] -
Lan, T.; McLaughlin, L. W. Biochemistry 2001, 40, 968–976. doi:10.1021/bi0015212
Return to citation in text: [1] -
Geyer, C. R.; Battersby, T. R.; Benner, S. A. Structure 2003, 11, 1485–1498. doi:10.1016/j.str.2003.11.008
Return to citation in text: [1] [2] [3] -
Benner, S. A. Acc. Chem. Res. 2004, 37, 784–797. doi:10.1021/ar040004z
Return to citation in text: [1] [2] -
Benner, S. A.; Sismour, A. M. Nat. Rev. Genet. 2005, 6, 533–543. doi:10.1038/nrg1637
Return to citation in text: [1] [2] -
Romeo, G.; Chiacchio, U.; Corsaro, A.; Merino, P. Chem. Rev. 2010, 110, 3337–3370. doi:10.1021/cr800464r
Return to citation in text: [1] [2] -
De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] -
Kool, E. T. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. doi:10.1146/annurev.biophys.30.1.1
Return to citation in text: [1] [2] -
Berti, P. J.; McCann, J. A. B. Chem. Rev. 2006, 106, 506–555. doi:10.1021/cr040461t
Return to citation in text: [1] [2] [3] [4] -
Ho, M.-C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P. C.; Evans, G. B.; Clinch, K.; Almo, S. C.; Schramm, V. L. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 4805–4812. doi:10.1073/pnas.0913439107
Return to citation in text: [1] [2] -
Malyshev, D. A.; Dhami, K.; Lavergne, T.; Chen, T.; Dai, N.; Foster, J. M.; Correa, I. R.; Romesberg, F. E. Nature 2014, 509, 385–388. doi:10.1038/nature13314
Return to citation in text: [1] -
Malyshev, D. A.; Romesberg, F. E. Angew. Chem., Int. Ed. 2015, 54, 11930–11944. doi:10.1002/anie.201502890
Return to citation in text: [1] -
Rios, A. C.; Yu, H. T.; Tor, Y. J. Phys. Org. Chem. 2015, 28, 173–180. doi:10.1002/poc.3318
Return to citation in text: [1] [2] -
Lenz, S. A. P.; Kohout, J. D.; Wetmore, S. D. J. Phys. Chem. B 2016, 120, 12795–12806. doi:10.1021/acs.jpcb.6b09620
Return to citation in text: [1] [2] [3] -
Wellington, K. W.; Benner, S. A. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 1309–1333. doi:10.1080/15257770600917013
Return to citation in text: [1] [2] [3] [4] -
Lindahl, T.; Karlstrom, O. Biochemistry 1973, 12, 5151–5154. doi:10.1021/bi00749a020
Return to citation in text: [1] [2] -
Lindahl, T.; Nyberg, B. Biochemistry 1974, 13, 3405–3410. doi:10.1021/bi00713a035
Return to citation in text: [1] [2] -
Lindahl, T. Nature 1993, 362, 709–715. doi:10.1038/362709a0
Return to citation in text: [1] [2] -
Levy, M.; Miller, S. L. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 7933–7938.
Return to citation in text: [1] [2] -
Schroeder, G. K.; Wolfenden, R. Biochemistry 2007, 46, 13638–13647. doi:10.1021/bi701480f
Return to citation in text: [1] -
Stockbridge, R. B.; Schroeder, G. K.; Wolfenden, R. Bioorg. Chem. 2010, 38, 224–228. doi:10.1016/j.bioorg.2010.05.003
Return to citation in text: [1] [2] -
Slater, A. G.; Hu, Y.; Yang, L.; Argent, S. P.; Lewis, W.; Blunt, M. O.; Champness, N. R. Chem. Sci. 2015, 6, 1562–1569. doi:10.1039/C4SC03531C
Return to citation in text: [1] -
Davis, F. F.; Allen, F. W. J. Biol. Chem. 1957, 227, 807–815.
Return to citation in text: [1] [2] [3] [4] -
Michelson, A. M.; Cohn, W. E. Biochemistry 1962, 1, 490–495. doi:10.1021/bi00909a020
Return to citation in text: [1] [2] [3] [4] -
Cortese, R.; Kammen, H. O.; Spengler, S. J.; Ames, B. N. J. Biol. Chem. 1974, 249, 1103–1108.
Return to citation in text: [1] [2] [3] [4] -
Daves, G. D., Jr.; Cheng, C. C. Prog. Med. Chem. 1976, 13, 303–349. doi:10.1016/S0079-6468(08)70141-3
Return to citation in text: [1] [2] [3] [4] -
Hacksell, U.; Daves, G. D., Jr. Prog. Med. Chem. 1985, 22, 1–65. doi:10.1016/S0079-6468(08)70228-5
Return to citation in text: [1] [2] [3] [4] -
Samuelsson, T.; Boren, T.; Johansen, T. I.; Lustig, F. J. Biol. Chem. 1988, 263, 13692–13699.
Return to citation in text: [1] [2] [3] [4] -
Townsend, L. B., Ed. Chemistry of Nucleosides and Nucleotides; 1994; Vol. 3.
Return to citation in text: [1] -
Maier, L.; Hylse, O.; Necas, M.; Trbusek, M.; Ytre-Arne, M.; Dalhus, B.; Bjoras, M.; Paruch, K. Tetrahedron Lett. 2014, 55, 3713–3716. doi:10.1016/j.tetlet.2014.05.030
Return to citation in text: [1] [2] [3] -
Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] -
Lim, M. I.; Ren, W. Y.; Otter, B. A.; Klein, R. S. J. Org. Chem. 1983, 48, 780–788. doi:10.1021/jo00154a005
Return to citation in text: [1] [2] -
Chu, M. Y.; Zuckerman, L. B.; Sato, S.; Crabtree, G. W.; Bogden, A. E.; Lim, M. I.; Klein, R. S. Biochem. Pharmacol. 1984, 33, 1229–1234. doi:10.1016/0006-2952(84)90174-6
Return to citation in text: [1] -
Joubert, N.; Pohl, R.; Klepetárova, B.; Hocek, M. J. Org. Chem. 2007, 72, 6797–6805. doi:10.1021/jo0709504
Return to citation in text: [1] -
Berger, M.; Ogawa, A. K.; McMinn, D. L.; Wu, Y.; Schultz, P. G.; Romesberg, F. E. Angew. Chem., Int. Ed. 2000, 39, 2940–2942. doi:10.1002/1521-3773(20000818)39:16<2940::AID-ANIE2940>3.0.CO;2-#
Return to citation in text: [1] -
Kim, H.-J.; Leal, N. A.; Hoshika, S.; Benner, S. A. J. Org. Chem. 2014, 79, 3194–3199. doi:10.1021/jo402665d
Return to citation in text: [1] [2] -
Teo, Y. N.; Kool, E. T. Chem. Rev. 2012, 112, 4221–4245. doi:10.1021/cr100351g
Return to citation in text: [1] -
Daves, G. D., Jr. Acc. Chem. Res. 1990, 23, 201–206. doi:10.1021/ar00174a006
Return to citation in text: [1] -
Krohn, K.; Heins, H.; Wielckens, K. J. Med. Chem. 1992, 35, 511–517. doi:10.1021/jm00081a012
Return to citation in text: [1] [2] -
Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] -
Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] [13] [14] -
Townsend, L. B., Ed. Chemistry of Nucleosides and Nucleotides; 1988; Vol. 1.
Return to citation in text: [1] -
Lim, M.-I.; Klein, R. S. Tetrahedron Lett. 1981, 22, 25–28. doi:10.1016/0040-4039(81)80031-7
Return to citation in text: [1] -
Ren, W. Y.; Lim, M. I.; Otter, B. A.; Klein, R. S. J. Org. Chem. 1982, 47, 4633–4637. doi:10.1021/jo00145a005
Return to citation in text: [1] -
Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] -
Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] -
Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] -
Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] -
Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] -
Evans, G. B.; Schramm, V. L.; Tyler, P. C. Curr. Med. Chem. 2015, 22, 3897–3909. doi:10.2174/0929867322666150821100851
Return to citation in text: [1] [2] -
Cho, A.; Zhang, L.; Xu, J.; Babusis, D.; Butler, T.; Lee, R.; Saunders, O. L.; Wang, T.; Parrish, J.; Perry, J.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 4127–4132. doi:10.1016/j.bmcl.2012.04.065
Return to citation in text: [1] [2] [3] -
Silveira, G. P.; Cardozo, H. M.; Rossa, T. A.; Sá, M. M. Curr. Org. Synth. 2015, 12, 584–602. doi:10.2174/157017941205150821130147
Return to citation in text: [1] [2] -
Batra, H.; Moriarty, R. M.; Penmasta, R.; Sharma, V.; Stanciuc, G.; Staszewski, J. P.; Tuladhar, S. M.; Walsh, D. A.; Datla, S.; Krishnaswamy, S. Org. Process Res. Dev. 2006, 10, 484–486. doi:10.1021/op050222n
Return to citation in text: [1] -
van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419
Return to citation in text: [1] [2] [3] [4] [5] [6] -
Matulic-Adamic, J.; Beigelman, L.; Portmann, S.; Egli, M.; Usman, N. J. Org. Chem. 1996, 61, 3909–3911. doi:10.1021/jo960091b
Return to citation in text: [1] [2] -
Czernecki, S.; Ville, G. J. Org. Chem. 1989, 54, 610–612. doi:10.1021/jo00264a020
Return to citation in text: [1] [2] -
Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Woerpel, K. A. J. Am. Chem. Soc. 1999, 121, 12208–12209. doi:10.1021/ja993349z
Return to citation in text: [1] [2] -
Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Smith, D. M.; Woerpel, K. A. J. Am. Chem. Soc. 2005, 127, 10879–10884. doi:10.1021/ja0524043
Return to citation in text: [1] [2] -
Patil, S. A.; Otter, B. A.; Klein, R. S. Tetrahedron Lett. 1994, 35, 5339–5342. doi:10.1016/S0040-4039(00)73494-0
Return to citation in text: [1] [2] [3] [4] -
Eldrup, A. B.; Allerson, C. R.; Bennett, C. F.; Bera, S.; Bhat, B.; Bhat, N.; Bosserman, M. R.; Brooks, J.; Burlein, C.; Carroll, S. S.; Cook, P. D.; Getty, K. L.; MacCoss, M.; McMasters, D. R.; Olsen, D. B.; Prakash, T. P.; Prhavc, M.; Song, Q.; Tomassini, J. E.; Xia, J. J. Med. Chem. 2004, 47, 2283–2295. doi:10.1021/jm030424e
Return to citation in text: [1] -
Shi, J.; Du, J.; Ma, T.; Pankiewicz, K. W.; Patterson, S. E.; Tharnish, P. M.; McBrayer, T. R.; Stuyver, L. J.; Otto, M. J.; Chu, C. K.; Schinazi, R. F.; Watanabe, K. A. Bioorg. Med. Chem. 2005, 13, 1641–1652. doi:10.1016/j.bmc.2004.12.011
Return to citation in text: [1] -
Wauchope, O. R.; Tomney, M. J.; Pepper, J. L.; Korba, B. E.; Seley-Radtke, K. L. Org. Lett. 2010, 12, 4466–4469. doi:10.1021/ol101482h
Return to citation in text: [1] -
Carroll, S. S.; Tomassini, J. E.; Bosserman, M.; Getty, K.; Stahlhut, M. W.; Eldrup, A. B.; Bhat, B.; Hall, D.; Simcoe, A. L.; LaFemina, R.; Rutkowski, C. A.; Wolanski, B.; Yang, Z.; Migliaccio, G.; De Francesco, R.; Kuo, L. C.; MacCoss, M.; Olsen, D. B. J. Biol. Chem. 2003, 278, 11979–11984. doi:10.1074/jbc.M210914200
Return to citation in text: [1] -
Migliaccio, G.; Tomassini, J. E.; Carroll, S. S.; Tomei, L.; Altamura, S.; Bhat, B.; Bartholomew, L.; Bosserman, M. R.; Ceccacci, A.; Colwell, L. F.; Cortese, R.; De Francesco, R.; Eldrup, A. B.; Getty, K. L.; Hou, X. S.; LaFemina, R. L.; Ludmerer, S. W.; MacCoss, M.; McMasters, D. R.; Stahlhut, M. W.; Olsen, D. B.; Hazuda, D. J.; Flores, O. A. J. Biol. Chem. 2003, 278, 49164–49170. doi:10.1074/jbc.M305041200
Return to citation in text: [1] -
Clark, J. L.; Hollecker, L.; Mason, J. C.; Stuyver, L. J.; Tharnish, P. M.; Lostia, S.; McBrayer, T. R.; Schinazi, R. F.; Watanabe, K. A.; Otto, M. J.; Furman, P. A.; Stec, W. J.; Patterson, S. E.; Pankiewicz, K. W. J. Med. Chem. 2005, 48, 5504–5508. doi:10.1021/jm0502788
Return to citation in text: [1] -
Sofia, M. J.; Bao, D.; Chang, W.; Du, J.; Nagarathnam, D.; Rachakonda, S.; Reddy, P. G.; Ross, B. S.; Wang, P.; Zhang, H.-R.; Bansal, S.; Espiritu, C.; Keilman, M.; Lam, A. M.; Steuer, H. M. M.; Niu, C.; Otto, M. J.; Furman, P. A. J. Med. Chem. 2010, 53, 7202–7218. doi:10.1021/jm100863x
Return to citation in text: [1] -
Chang, W.; Bao, D.; Chun, B.-K.; Naduthambi, D.; Nagarathnam, D.; Rachakonda, S.; Reddy, P. G.; Ross, B. S.; Zhang, H.-R.; Bansal, S.; Espiritu, C. L.; Keilman, M.; Lam, A. M.; Niu, C.; Steuer, H. M.; Furman, P. A.; Otto, M. J.; Sofia, M. J. ACS Med. Chem. Lett. 2011, 2, 130–135. doi:10.1021/ml100209f
Return to citation in text: [1] -
Simonovich, S. P.; Van Humbeck, J. F.; MacMillan, D. W. C. Chem. Sci. 2012, 3, 58–61. doi:10.1039/C1SC00556A
Return to citation in text: [1] -
Warren, T. K.; Wells, J.; Panchal, R. G.; Stuthman, K. S.; Garza, N. L.; Van Tongeren, S. A.; Dong, L.; Retterer, C. J.; Eaton, B. P.; Pegoraro, G.; Honnold, S.; Bantia, S.; Kotian, P.; Chen, X.; Taubenheim, B. R.; Welch, L. S.; Minning, D. M.; Babu, Y. S.; Sheridan, W. P.; Bavari, S. Nature 2014, 508, 402–405. doi:10.1038/nature13027
Return to citation in text: [1] [2] -
Bergeron-Brlek, M.; Meanwell, M.; Britton, R. Nat. Commun. 2015, 6, 6903. doi:10.1038/ncomms7903
Return to citation in text: [1] -
Blauenburg, B.; Oja, T.; Klika, K. D.; Metsä-Ketelä, M. ACS Chem. Biol. 2013, 8, 2377–2382. doi:10.1021/cb400384c
Return to citation in text: [1] -
Thapa, K.; Oja, T.; Metsae-Ketelae, M. FEBS J. 2014, 281, 4439–4449. doi:10.1111/febs.12950
Return to citation in text: [1] -
Carlile, T. M.; Rojas-Duran, M. F.; Zinshteyn, B.; Shin, H.; Bartoli, K. M.; Gilbert, W. V. Nature 2014, 515, 143–146. doi:10.1038/nature13802
Return to citation in text: [1] -
Friedt, J.; Leavens, F. M. V.; Mercier, E.; Wieden, H.-J.; Kothe, U. Nucleic Acids Res. 2014, 42, 3857–3870. doi:10.1093/nar/gkt1331
Return to citation in text: [1] -
Veerareddygari, G. R.; Singh, S. K.; Mueller, E. G. J. Am. Chem. Soc. 2016, 138, 7852–7855. doi:10.1021/jacs.6b04491
Return to citation in text: [1] -
Palmu, K.; Rosenqvist, P.; Thapa, K.; Ilina, Y.; Siitonen, V.; Baral, B.; Makinen, J.; Belogurov, G.; Virta, P.; Niemi, J.; Metsä-Ketelä, M. ACS Chem. Biol. 2017, 12, 1472–1477. doi:10.1021/acschembio.7b00078
Return to citation in text: [1] -
Ko, Y.; Wang, S.-A.; Ogasawara, Y.; Ruszczycky, M. W.; Liu, H.-w. Org. Lett. 2017, 19, 1426–1429. doi:10.1021/acs.orglett.7b00355
Return to citation in text: [1]
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 82. | Matulic-Adamic, J.; Beigelman, L.; Portmann, S.; Egli, M.; Usman, N. J. Org. Chem. 1996, 61, 3909–3911. doi:10.1021/jo960091b |
| 83. | Czernecki, S.; Ville, G. J. Org. Chem. 1989, 54, 610–612. doi:10.1021/jo00264a020 |
| 82. | Matulic-Adamic, J.; Beigelman, L.; Portmann, S.; Egli, M.; Usman, N. J. Org. Chem. 1996, 61, 3909–3911. doi:10.1021/jo960091b |
| 83. | Czernecki, S.; Ville, G. J. Org. Chem. 1989, 54, 610–612. doi:10.1021/jo00264a020 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 95. | Simonovich, S. P.; Van Humbeck, J. F.; MacMillan, D. W. C. Chem. Sci. 2012, 3, 58–61. doi:10.1039/C1SC00556A |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 77. | Cho, A.; Zhang, L.; Xu, J.; Babusis, D.; Butler, T.; Lee, R.; Saunders, O. L.; Wang, T.; Parrish, J.; Perry, J.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 4127–4132. doi:10.1016/j.bmcl.2012.04.065 |
| 86. | Patil, S. A.; Otter, B. A.; Klein, R. S. Tetrahedron Lett. 1994, 35, 5339–5342. doi:10.1016/S0040-4039(00)73494-0 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 52. | Maier, L.; Hylse, O.; Necas, M.; Trbusek, M.; Ytre-Arne, M.; Dalhus, B.; Bjoras, M.; Paruch, K. Tetrahedron Lett. 2014, 55, 3713–3716. doi:10.1016/j.tetlet.2014.05.030 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 84. | Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Woerpel, K. A. J. Am. Chem. Soc. 1999, 121, 12208–12209. doi:10.1021/ja993349z |
| 85. | Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Smith, D. M.; Woerpel, K. A. J. Am. Chem. Soc. 2005, 127, 10879–10884. doi:10.1021/ja0524043 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 18. | Maffioli, S. I.; Zhang, Y.; Degen, D.; Carzaniga, T.; Del Gatto, G.; Serina, S.; Monciardini, P.; Mazzetti, C.; Guglierame, P.; Candiani, G.; Chiriac, A. I.; Facchetti, G.; Kaltofen, P.; Sahl, H.-G.; Dehò, G.; Donadio, S.; Ebright, R. H. Cell 2017, 169, 1240–1248. doi:10.1016/j.cell.2017.05.042 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 96. | Warren, T. K.; Wells, J.; Panchal, R. G.; Stuthman, K. S.; Garza, N. L.; Van Tongeren, S. A.; Dong, L.; Retterer, C. J.; Eaton, B. P.; Pegoraro, G.; Honnold, S.; Bantia, S.; Kotian, P.; Chen, X.; Taubenheim, B. R.; Welch, L. S.; Minning, D. M.; Babu, Y. S.; Sheridan, W. P.; Bavari, S. Nature 2014, 508, 402–405. doi:10.1038/nature13027 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 86. | Patil, S. A.; Otter, B. A.; Klein, R. S. Tetrahedron Lett. 1994, 35, 5339–5342. doi:10.1016/S0040-4039(00)73494-0 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 77. | Cho, A.; Zhang, L.; Xu, J.; Babusis, D.; Butler, T.; Lee, R.; Saunders, O. L.; Wang, T.; Parrish, J.; Perry, J.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 4127–4132. doi:10.1016/j.bmcl.2012.04.065 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 86. | Patil, S. A.; Otter, B. A.; Klein, R. S. Tetrahedron Lett. 1994, 35, 5339–5342. doi:10.1016/S0040-4039(00)73494-0 |
| 80. | van Rijssel, E. R.; van Delft, P.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. Angew. Chem., Int. Ed. 2014, 53, 10381–10385. doi:10.1002/anie.201405477 |
| 81. | van Rijssel, E. R.; van Delft, P.; van Marle, D. V.; Bijvoets, S. M.; Lodder, G.; Overkleeft, H. S.; van der Marel, G. A.; Filippov, D. V.; Codée, J. D. C. J. Org. Chem. 2015, 80, 4553–4565. doi:10.1021/acs.joc.5b00419 |
| 84. | Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Woerpel, K. A. J. Am. Chem. Soc. 1999, 121, 12208–12209. doi:10.1021/ja993349z |
| 85. | Larsen, C. H.; Ridgway, B. H.; Shaw, J. T.; Smith, D. M.; Woerpel, K. A. J. Am. Chem. Soc. 2005, 127, 10879–10884. doi:10.1021/ja0524043 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 98. | Blauenburg, B.; Oja, T.; Klika, K. D.; Metsä-Ketelä, M. ACS Chem. Biol. 2013, 8, 2377–2382. doi:10.1021/cb400384c |
| 99. | Thapa, K.; Oja, T.; Metsae-Ketelae, M. FEBS J. 2014, 281, 4439–4449. doi:10.1111/febs.12950 |
| 100. | Carlile, T. M.; Rojas-Duran, M. F.; Zinshteyn, B.; Shin, H.; Bartoli, K. M.; Gilbert, W. V. Nature 2014, 515, 143–146. doi:10.1038/nature13802 |
| 101. | Friedt, J.; Leavens, F. M. V.; Mercier, E.; Wieden, H.-J.; Kothe, U. Nucleic Acids Res. 2014, 42, 3857–3870. doi:10.1093/nar/gkt1331 |
| 102. | Veerareddygari, G. R.; Singh, S. K.; Mueller, E. G. J. Am. Chem. Soc. 2016, 138, 7852–7855. doi:10.1021/jacs.6b04491 |
| 103. | Palmu, K.; Rosenqvist, P.; Thapa, K.; Ilina, Y.; Siitonen, V.; Baral, B.; Makinen, J.; Belogurov, G.; Virta, P.; Niemi, J.; Metsä-Ketelä, M. ACS Chem. Biol. 2017, 12, 1472–1477. doi:10.1021/acschembio.7b00078 |
| 104. | Ko, Y.; Wang, S.-A.; Ogasawara, Y.; Ruszczycky, M. W.; Liu, H.-w. Org. Lett. 2017, 19, 1426–1429. doi:10.1021/acs.orglett.7b00355 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 76. | Evans, G. B.; Schramm, V. L.; Tyler, P. C. Curr. Med. Chem. 2015, 22, 3897–3909. doi:10.2174/0929867322666150821100851 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 97. | Bergeron-Brlek, M.; Meanwell, M.; Britton, R. Nat. Commun. 2015, 6, 6903. doi:10.1038/ncomms7903 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 1. | Miescher, F. Med. Chem. Unters. 1871, 441–460. |
| 2. | Avery, O. T.; Macleod, C. M.; McCarty, M. J. Exp. Med. 1944, 79, 137–158. doi:10.1084/jem.79.2.137 |
| 3. | Chargaff, E.; Lipshitz, R. J. Am. Chem. Soc. 1953, 75, 3658–3661. doi:10.1021/ja01111a016 |
| 4. | Franklin, R. E.; Gosling, R. G. Nature 1953, 172, 156–157. doi:10.1038/172156a0 |
| 5. | Watson, J. D.; Crick, F. H. C. Nature 1953, 171, 737–738. doi:10.1038/171737a0 |
| 6. | Watson, J. D.; Crick, F. H. C. Nature 1953, 171, 964–967. doi:10.1038/171964b0 |
| 7. | Nelson, D. L.; Cox, M. M. Lehninger Principles of Biochemistry, 4th ed.; Freemand and Company, 2005. |
| 8. | Matera, A. G.; Terns, R. M.; Terns, M. P. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. doi:10.1038/nrm2124 |
| 9. | Rhodes, D.; Lipps, H. J. Nucleic Acids Res. 2015, 43, 8627–8637. doi:10.1093/nar/gkv862 |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 86. | Patil, S. A.; Otter, B. A.; Klein, R. S. Tetrahedron Lett. 1994, 35, 5339–5342. doi:10.1016/S0040-4039(00)73494-0 |
| 96. | Warren, T. K.; Wells, J.; Panchal, R. G.; Stuthman, K. S.; Garza, N. L.; Van Tongeren, S. A.; Dong, L.; Retterer, C. J.; Eaton, B. P.; Pegoraro, G.; Honnold, S.; Bantia, S.; Kotian, P.; Chen, X.; Taubenheim, B. R.; Welch, L. S.; Minning, D. M.; Babu, Y. S.; Sheridan, W. P.; Bavari, S. Nature 2014, 508, 402–405. doi:10.1038/nature13027 |
| 23. | Cihak, A. Oncology 1974, 30, 405–422. doi:10.1159/000224981 |
| 24. | Lan, T.; McLaughlin, L. W. Biochemistry 2001, 40, 968–976. doi:10.1021/bi0015212 |
| 25. | Geyer, C. R.; Battersby, T. R.; Benner, S. A. Structure 2003, 11, 1485–1498. doi:10.1016/j.str.2003.11.008 |
| 26. | Benner, S. A. Acc. Chem. Res. 2004, 37, 784–797. doi:10.1021/ar040004z |
| 27. | Benner, S. A.; Sismour, A. M. Nat. Rev. Genet. 2005, 6, 533–543. doi:10.1038/nrg1637 |
| 28. | Romeo, G.; Chiacchio, U.; Corsaro, A.; Merino, P. Chem. Rev. 2010, 110, 3337–3370. doi:10.1021/cr800464r |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 30. | Kool, E. T. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. doi:10.1146/annurev.biophys.30.1.1 |
| 45. | Davis, F. F.; Allen, F. W. J. Biol. Chem. 1957, 227, 807–815. |
| 46. | Michelson, A. M.; Cohn, W. E. Biochemistry 1962, 1, 490–495. doi:10.1021/bi00909a020 |
| 47. | Cortese, R.; Kammen, H. O.; Spengler, S. J.; Ames, B. N. J. Biol. Chem. 1974, 249, 1103–1108. |
| 50. | Samuelsson, T.; Boren, T.; Johansen, T. I.; Lustig, F. J. Biol. Chem. 1988, 263, 13692–13699. |
| 54. | Lim, M. I.; Ren, W. Y.; Otter, B. A.; Klein, R. S. J. Org. Chem. 1983, 48, 780–788. doi:10.1021/jo00154a005 |
| 55. | Chu, M. Y.; Zuckerman, L. B.; Sato, S.; Crabtree, G. W.; Bogden, A. E.; Lim, M. I.; Klein, R. S. Biochem. Pharmacol. 1984, 33, 1229–1234. doi:10.1016/0006-2952(84)90174-6 |
| 56. | Joubert, N.; Pohl, R.; Klepetárova, B.; Hocek, M. J. Org. Chem. 2007, 72, 6797–6805. doi:10.1021/jo0709504 |
| 57. | Berger, M.; Ogawa, A. K.; McMinn, D. L.; Wu, Y.; Schultz, P. G.; Romesberg, F. E. Angew. Chem., Int. Ed. 2000, 39, 2940–2942. doi:10.1002/1521-3773(20000818)39:16<2940::AID-ANIE2940>3.0.CO;2-# |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 13. | Jordheim, L. P.; Durantel, D.; Zoulim, F.; Dumontet, C. Nat. Rev. Drug Discovery 2013, 12, 447–464. doi:10.1038/nrd4010 |
| 14. | Coats, S. J.; Garnier-Amblard, E. C.; Amblard, F.; Ehteshami, M.; Amiralaei, S.; Zhang, H.; Zhou, L.; Boucle, S. R. L.; Lu, X.; Bondada, L.; Shelton, J. R.; Li, H.; Liu, P.; Li, C.; Cho, J. H.; Chavre, S. N.; Zhou, S.; Mathew, J.; Schinazi, R. F. Antiviral Res. 2014, 102, 119–147. doi:10.1016/j.antiviral.2013.11.008 |
| 15. | Pradere, U.; Garnier-Amblard, E. C.; Coats, S. J.; Amblard, F.; Schinazi, R. F. Chem. Rev. 2014, 114, 9154–9218. doi:10.1021/cr5002035 |
| 25. | Geyer, C. R.; Battersby, T. R.; Benner, S. A. Structure 2003, 11, 1485–1498. doi:10.1016/j.str.2003.11.008 |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 37. | Wellington, K. W.; Benner, S. A. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 1309–1333. doi:10.1080/15257770600917013 |
| 58. | Kim, H.-J.; Leal, N. A.; Hoshika, S.; Benner, S. A. J. Org. Chem. 2014, 79, 3194–3199. doi:10.1021/jo402665d |
| 7. | Nelson, D. L.; Cox, M. M. Lehninger Principles of Biochemistry, 4th ed.; Freemand and Company, 2005. |
| 45. | Davis, F. F.; Allen, F. W. J. Biol. Chem. 1957, 227, 807–815. |
| 46. | Michelson, A. M.; Cohn, W. E. Biochemistry 1962, 1, 490–495. doi:10.1021/bi00909a020 |
| 47. | Cortese, R.; Kammen, H. O.; Spengler, S. J.; Ames, B. N. J. Biol. Chem. 1974, 249, 1103–1108. |
| 48. | Daves, G. D., Jr.; Cheng, C. C. Prog. Med. Chem. 1976, 13, 303–349. doi:10.1016/S0079-6468(08)70141-3 |
| 49. | Hacksell, U.; Daves, G. D., Jr. Prog. Med. Chem. 1985, 22, 1–65. doi:10.1016/S0079-6468(08)70228-5 |
| 50. | Samuelsson, T.; Boren, T.; Johansen, T. I.; Lustig, F. J. Biol. Chem. 1988, 263, 13692–13699. |
| 51. | Townsend, L. B., Ed. Chemistry of Nucleosides and Nucleotides; 1994; Vol. 3. |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 10. | Hamma, T.; Ferre-D'Amare, A. R. Chem. Biol. 2006, 13, 1125–1135. doi:10.1016/j.chembiol.2006.09.009 |
| 11. | McDonald, M. K.; Miracco, E. J.; Chen, J.; Xie, Y.; Mueller, E. G. Biochemistry 2011, 50, 426–436. doi:10.1021/bi101737z |
| 12. | Boettcher, T.; Sieber, S. A. J. Am. Chem. Soc. 2010, 132, 6964–6972. doi:10.1021/ja909150y |
| 13. | Jordheim, L. P.; Durantel, D.; Zoulim, F.; Dumontet, C. Nat. Rev. Drug Discovery 2013, 12, 447–464. doi:10.1038/nrd4010 |
| 14. | Coats, S. J.; Garnier-Amblard, E. C.; Amblard, F.; Ehteshami, M.; Amiralaei, S.; Zhang, H.; Zhou, L.; Boucle, S. R. L.; Lu, X.; Bondada, L.; Shelton, J. R.; Li, H.; Liu, P.; Li, C.; Cho, J. H.; Chavre, S. N.; Zhou, S.; Mathew, J.; Schinazi, R. F. Antiviral Res. 2014, 102, 119–147. doi:10.1016/j.antiviral.2013.11.008 |
| 15. | Pradere, U.; Garnier-Amblard, E. C.; Coats, S. J.; Amblard, F.; Schinazi, R. F. Chem. Rev. 2014, 114, 9154–9218. doi:10.1021/cr5002035 |
| 16. | Wiegmann, D.; Koppermann, S.; Wirth, M.; Niro, G.; Leyerer, K.; Ducho, C. Beilstein J. Org. Chem. 2016, 12, 769–795. doi:10.3762/bjoc.12.77 |
| 17. | Serpi, M.; Ferrari, V.; Pertusati, F. J. Med. Chem. 2016, 59, 10343–10382. doi:10.1021/acs.jmedchem.6b00325 |
| 18. | Maffioli, S. I.; Zhang, Y.; Degen, D.; Carzaniga, T.; Del Gatto, G.; Serina, S.; Monciardini, P.; Mazzetti, C.; Guglierame, P.; Candiani, G.; Chiriac, A. I.; Facchetti, G.; Kaltofen, P.; Sahl, H.-G.; Dehò, G.; Donadio, S.; Ebright, R. H. Cell 2017, 169, 1240–1248. doi:10.1016/j.cell.2017.05.042 |
| 19. | Davis, J. T.; Spada, G. P. Chem. Soc. Rev. 2007, 36, 296–313. doi:10.1039/B600282J |
| 20. | Jones, M. R.; Seeman, N. C.; Mirkin, C. A. Science 2015, 347, 1260901. doi:10.1126/science.1260901 |
| 21. | Krishnan, Y.; Simmel, F. C. Angew. Chem., Int. Ed. 2011, 50, 3124–3156. doi:10.1002/anie.200907223 |
| 22. | Fenniri, H.; Temburnikar, K. W.; Johnson, R. S. In Comprehensive Supramolecular Chemistry II; Atwood, J. L., Ed.; Elsevier: Oxford, 2017; pp 83–113. doi:10.1016/B978-0-12-409547-2.12583-1 |
| 52. | Maier, L.; Hylse, O.; Necas, M.; Trbusek, M.; Ytre-Arne, M.; Dalhus, B.; Bjoras, M.; Paruch, K. Tetrahedron Lett. 2014, 55, 3713–3716. doi:10.1016/j.tetlet.2014.05.030 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 38. | Lindahl, T.; Karlstrom, O. Biochemistry 1973, 12, 5151–5154. doi:10.1021/bi00749a020 |
| 39. | Lindahl, T.; Nyberg, B. Biochemistry 1974, 13, 3405–3410. doi:10.1021/bi00713a035 |
| 40. | Lindahl, T. Nature 1993, 362, 709–715. doi:10.1038/362709a0 |
| 41. | Levy, M.; Miller, S. L. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 7933–7938. |
| 43. | Stockbridge, R. B.; Schroeder, G. K.; Wolfenden, R. Bioorg. Chem. 2010, 38, 224–228. doi:10.1016/j.bioorg.2010.05.003 |
| 31. | Berti, P. J.; McCann, J. A. B. Chem. Rev. 2006, 106, 506–555. doi:10.1021/cr040461t |
| 36. | Lenz, S. A. P.; Kohout, J. D.; Wetmore, S. D. J. Phys. Chem. B 2016, 120, 12795–12806. doi:10.1021/acs.jpcb.6b09620 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 38. | Lindahl, T.; Karlstrom, O. Biochemistry 1973, 12, 5151–5154. doi:10.1021/bi00749a020 |
| 39. | Lindahl, T.; Nyberg, B. Biochemistry 1974, 13, 3405–3410. doi:10.1021/bi00713a035 |
| 40. | Lindahl, T. Nature 1993, 362, 709–715. doi:10.1038/362709a0 |
| 41. | Levy, M.; Miller, S. L. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 7933–7938. |
| 42. | Schroeder, G. K.; Wolfenden, R. Biochemistry 2007, 46, 13638–13647. doi:10.1021/bi701480f |
| 43. | Stockbridge, R. B.; Schroeder, G. K.; Wolfenden, R. Bioorg. Chem. 2010, 38, 224–228. doi:10.1016/j.bioorg.2010.05.003 |
| 44. | Slater, A. G.; Hu, Y.; Yang, L.; Argent, S. P.; Lewis, W.; Blunt, M. O.; Champness, N. R. Chem. Sci. 2015, 6, 1562–1569. doi:10.1039/C4SC03531C |
| 31. | Berti, P. J.; McCann, J. A. B. Chem. Rev. 2006, 106, 506–555. doi:10.1021/cr040461t |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 37. | Wellington, K. W.; Benner, S. A. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 1309–1333. doi:10.1080/15257770600917013 |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 25. | Geyer, C. R.; Battersby, T. R.; Benner, S. A. Structure 2003, 11, 1485–1498. doi:10.1016/j.str.2003.11.008 |
| 26. | Benner, S. A. Acc. Chem. Res. 2004, 37, 784–797. doi:10.1021/ar040004z |
| 27. | Benner, S. A.; Sismour, A. M. Nat. Rev. Genet. 2005, 6, 533–543. doi:10.1038/nrg1637 |
| 28. | Romeo, G.; Chiacchio, U.; Corsaro, A.; Merino, P. Chem. Rev. 2010, 110, 3337–3370. doi:10.1021/cr800464r |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 30. | Kool, E. T. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. doi:10.1146/annurev.biophys.30.1.1 |
| 31. | Berti, P. J.; McCann, J. A. B. Chem. Rev. 2006, 106, 506–555. doi:10.1021/cr040461t |
| 32. | Ho, M.-C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P. C.; Evans, G. B.; Clinch, K.; Almo, S. C.; Schramm, V. L. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 4805–4812. doi:10.1073/pnas.0913439107 |
| 33. | Malyshev, D. A.; Dhami, K.; Lavergne, T.; Chen, T.; Dai, N.; Foster, J. M.; Correa, I. R.; Romesberg, F. E. Nature 2014, 509, 385–388. doi:10.1038/nature13314 |
| 34. | Malyshev, D. A.; Romesberg, F. E. Angew. Chem., Int. Ed. 2015, 54, 11930–11944. doi:10.1002/anie.201502890 |
| 35. | Rios, A. C.; Yu, H. T.; Tor, Y. J. Phys. Org. Chem. 2015, 28, 173–180. doi:10.1002/poc.3318 |
| 36. | Lenz, S. A. P.; Kohout, J. D.; Wetmore, S. D. J. Phys. Chem. B 2016, 120, 12795–12806. doi:10.1021/acs.jpcb.6b09620 |
| 37. | Wellington, K. W.; Benner, S. A. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 1309–1333. doi:10.1080/15257770600917013 |
| 31. | Berti, P. J.; McCann, J. A. B. Chem. Rev. 2006, 106, 506–555. doi:10.1021/cr040461t |
| 35. | Rios, A. C.; Yu, H. T.; Tor, Y. J. Phys. Org. Chem. 2015, 28, 173–180. doi:10.1002/poc.3318 |
| 36. | Lenz, S. A. P.; Kohout, J. D.; Wetmore, S. D. J. Phys. Chem. B 2016, 120, 12795–12806. doi:10.1021/acs.jpcb.6b09620 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 18. | Maffioli, S. I.; Zhang, Y.; Degen, D.; Carzaniga, T.; Del Gatto, G.; Serina, S.; Monciardini, P.; Mazzetti, C.; Guglierame, P.; Candiani, G.; Chiriac, A. I.; Facchetti, G.; Kaltofen, P.; Sahl, H.-G.; Dehò, G.; Donadio, S.; Ebright, R. H. Cell 2017, 169, 1240–1248. doi:10.1016/j.cell.2017.05.042 |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 37. | Wellington, K. W.; Benner, S. A. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 1309–1333. doi:10.1080/15257770600917013 |
| 45. | Davis, F. F.; Allen, F. W. J. Biol. Chem. 1957, 227, 807–815. |
| 46. | Michelson, A. M.; Cohn, W. E. Biochemistry 1962, 1, 490–495. doi:10.1021/bi00909a020 |
| 47. | Cortese, R.; Kammen, H. O.; Spengler, S. J.; Ames, B. N. J. Biol. Chem. 1974, 249, 1103–1108. |
| 48. | Daves, G. D., Jr.; Cheng, C. C. Prog. Med. Chem. 1976, 13, 303–349. doi:10.1016/S0079-6468(08)70141-3 |
| 49. | Hacksell, U.; Daves, G. D., Jr. Prog. Med. Chem. 1985, 22, 1–65. doi:10.1016/S0079-6468(08)70228-5 |
| 50. | Samuelsson, T.; Boren, T.; Johansen, T. I.; Lustig, F. J. Biol. Chem. 1988, 263, 13692–13699. |
| 60. | Daves, G. D., Jr. Acc. Chem. Res. 1990, 23, 201–206. doi:10.1021/ar00174a006 |
| 61. | Krohn, K.; Heins, H.; Wielckens, K. J. Med. Chem. 1992, 35, 511–517. doi:10.1021/jm00081a012 |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 12. | Boettcher, T.; Sieber, S. A. J. Am. Chem. Soc. 2010, 132, 6964–6972. doi:10.1021/ja909150y |
| 29. | De Clercq, E. J. Med. Chem. 2016, 59, 2301–2311. doi:10.1021/acs.jmedchem.5b01157 |
| 59. | Teo, Y. N.; Kool, E. T. Chem. Rev. 2012, 112, 4221–4245. doi:10.1021/cr100351g |
| 45. | Davis, F. F.; Allen, F. W. J. Biol. Chem. 1957, 227, 807–815. |
| 46. | Michelson, A. M.; Cohn, W. E. Biochemistry 1962, 1, 490–495. doi:10.1021/bi00909a020 |
| 47. | Cortese, R.; Kammen, H. O.; Spengler, S. J.; Ames, B. N. J. Biol. Chem. 1974, 249, 1103–1108. |
| 50. | Samuelsson, T.; Boren, T.; Johansen, T. I.; Lustig, F. J. Biol. Chem. 1988, 263, 13692–13699. |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 61. | Krohn, K.; Heins, H.; Wielckens, K. J. Med. Chem. 1992, 35, 511–517. doi:10.1021/jm00081a012 |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 78. | Silveira, G. P.; Cardozo, H. M.; Rossa, T. A.; Sá, M. M. Curr. Org. Synth. 2015, 12, 584–602. doi:10.2174/157017941205150821130147 |
| 78. | Silveira, G. P.; Cardozo, H. M.; Rossa, T. A.; Sá, M. M. Curr. Org. Synth. 2015, 12, 584–602. doi:10.2174/157017941205150821130147 |
| 79. | Batra, H.; Moriarty, R. M.; Penmasta, R.; Sharma, V.; Stanciuc, G.; Staszewski, J. P.; Tuladhar, S. M.; Walsh, D. A.; Datla, S.; Krishnaswamy, S. Org. Process Res. Dev. 2006, 10, 484–486. doi:10.1021/op050222n |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 52. | Maier, L.; Hylse, O.; Necas, M.; Trbusek, M.; Ytre-Arne, M.; Dalhus, B.; Bjoras, M.; Paruch, K. Tetrahedron Lett. 2014, 55, 3713–3716. doi:10.1016/j.tetlet.2014.05.030 |
| 53. | Maier, L.; Khirsariya, P.; Hylse, O.; Adla, S. K.; Černová, L.; Poljak, M.; Krajčovičová, S.; Weis, E.; Drapelá, S.; Souček, K.; Paruch, K. J. Org. Chem. 2017, 82, 3382–3402. doi:10.1021/acs.joc.6b02594 |
| 58. | Kim, H.-J.; Leal, N. A.; Hoshika, S.; Benner, S. A. J. Org. Chem. 2014, 79, 3194–3199. doi:10.1021/jo402665d |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 69. | Cho, A.; Saunders, O. L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J. E.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 2705–2707. doi:10.1016/j.bmcl.2012.02.105 |
| 70. | Metobo, S. E.; Xu, J.; Saunders, O. L.; Butler, T.; Aktoudianakis, E.; Cho, A.; Kim, C. U. Tetrahedron Lett. 2012, 53, 484–486. doi:10.1016/j.tetlet.2011.11.055 |
| 71. | Draffan, A. G.; Frey, B.; Pool, B.; Gannon, C.; Tyndall, E. M.; Lilly, M.; Francom, P.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Nguyen, V. T. T.; Jeynes, T. P.; Wirth, V.; Luttick, A.; Tilmanis, D.; Thomas, J. D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Kukolj, G.; Simoneau, B.; McKercher, G.; Lagace, L.; Amad, M. a.; Bethell, R. C.; Tucker, S. P. ACS Med. Chem. Lett. 2014, 5, 679–684. doi:10.1021/ml500077j |
| 72. | Draffan, A. G.; Frey, B.; Fraser, B. H.; Pool, B.; Gannon, C.; Tyndall, E. M.; Cianci, J.; Harding, M.; Lilly, M.; Hufton, R.; Halim, R.; Jahangiri, S.; Bond, S.; Jeynes, T. P.; Nguyen, V. T. T.; Wirth, V.; Luttick, A.; Tilmanis, D.; Pryor, M.; Porter, K.; Morton, C. J.; Lin, B.; Duan, J.; Bethell, R. C.; Kukolj, G.; Simoneau, B.; Tucker, S. P. Bioorg. Med. Chem. Lett. 2014, 24, 4984–4988. doi:10.1016/j.bmcl.2014.09.030 |
| 73. | Dang, Q.; Zhang, Z.; Chen, T.; Tang, B.; He, X.; He, S.; Song, Y.; Bogen, S.; Girijavallabhan, V.; Olsen, D. B.; Meinke, P. T. Tetrahedron Lett. 2014, 55, 5092–5095. doi:10.1016/j.tetlet.2014.07.037 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 75. | Peifer, M.; Berger, R.; Shurtleff, V. W.; Conrad, J. C.; MacMillan, D. W. C. J. Am. Chem. Soc. 2014, 136, 5900–5903. doi:10.1021/ja502205q |
| 76. | Evans, G. B.; Schramm, V. L.; Tyler, P. C. Curr. Med. Chem. 2015, 22, 3897–3909. doi:10.2174/0929867322666150821100851 |
| 77. | Cho, A.; Zhang, L.; Xu, J.; Babusis, D.; Butler, T.; Lee, R.; Saunders, O. L.; Wang, T.; Parrish, J.; Perry, J.; Feng, J. Y.; Ray, A. S.; Kim, C. U. Bioorg. Med. Chem. Lett. 2012, 22, 4127–4132. doi:10.1016/j.bmcl.2012.04.065 |
| 87. | Eldrup, A. B.; Allerson, C. R.; Bennett, C. F.; Bera, S.; Bhat, B.; Bhat, N.; Bosserman, M. R.; Brooks, J.; Burlein, C.; Carroll, S. S.; Cook, P. D.; Getty, K. L.; MacCoss, M.; McMasters, D. R.; Olsen, D. B.; Prakash, T. P.; Prhavc, M.; Song, Q.; Tomassini, J. E.; Xia, J. J. Med. Chem. 2004, 47, 2283–2295. doi:10.1021/jm030424e |
| 88. | Shi, J.; Du, J.; Ma, T.; Pankiewicz, K. W.; Patterson, S. E.; Tharnish, P. M.; McBrayer, T. R.; Stuyver, L. J.; Otto, M. J.; Chu, C. K.; Schinazi, R. F.; Watanabe, K. A. Bioorg. Med. Chem. 2005, 13, 1641–1652. doi:10.1016/j.bmc.2004.12.011 |
| 89. | Wauchope, O. R.; Tomney, M. J.; Pepper, J. L.; Korba, B. E.; Seley-Radtke, K. L. Org. Lett. 2010, 12, 4466–4469. doi:10.1021/ol101482h |
| 90. | Carroll, S. S.; Tomassini, J. E.; Bosserman, M.; Getty, K.; Stahlhut, M. W.; Eldrup, A. B.; Bhat, B.; Hall, D.; Simcoe, A. L.; LaFemina, R.; Rutkowski, C. A.; Wolanski, B.; Yang, Z.; Migliaccio, G.; De Francesco, R.; Kuo, L. C.; MacCoss, M.; Olsen, D. B. J. Biol. Chem. 2003, 278, 11979–11984. doi:10.1074/jbc.M210914200 |
| 91. | Migliaccio, G.; Tomassini, J. E.; Carroll, S. S.; Tomei, L.; Altamura, S.; Bhat, B.; Bartholomew, L.; Bosserman, M. R.; Ceccacci, A.; Colwell, L. F.; Cortese, R.; De Francesco, R.; Eldrup, A. B.; Getty, K. L.; Hou, X. S.; LaFemina, R. L.; Ludmerer, S. W.; MacCoss, M.; McMasters, D. R.; Stahlhut, M. W.; Olsen, D. B.; Hazuda, D. J.; Flores, O. A. J. Biol. Chem. 2003, 278, 49164–49170. doi:10.1074/jbc.M305041200 |
| 92. | Clark, J. L.; Hollecker, L.; Mason, J. C.; Stuyver, L. J.; Tharnish, P. M.; Lostia, S.; McBrayer, T. R.; Schinazi, R. F.; Watanabe, K. A.; Otto, M. J.; Furman, P. A.; Stec, W. J.; Patterson, S. E.; Pankiewicz, K. W. J. Med. Chem. 2005, 48, 5504–5508. doi:10.1021/jm0502788 |
| 93. | Sofia, M. J.; Bao, D.; Chang, W.; Du, J.; Nagarathnam, D.; Rachakonda, S.; Reddy, P. G.; Ross, B. S.; Wang, P.; Zhang, H.-R.; Bansal, S.; Espiritu, C.; Keilman, M.; Lam, A. M.; Steuer, H. M. M.; Niu, C.; Otto, M. J.; Furman, P. A. J. Med. Chem. 2010, 53, 7202–7218. doi:10.1021/jm100863x |
| 94. | Chang, W.; Bao, D.; Chun, B.-K.; Naduthambi, D.; Nagarathnam, D.; Rachakonda, S.; Reddy, P. G.; Ross, B. S.; Zhang, H.-R.; Bansal, S.; Espiritu, C. L.; Keilman, M.; Lam, A. M.; Niu, C.; Steuer, H. M.; Furman, P. A.; Otto, M. J.; Sofia, M. J. ACS Med. Chem. Lett. 2011, 2, 130–135. doi:10.1021/ml100209f |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 48. | Daves, G. D., Jr.; Cheng, C. C. Prog. Med. Chem. 1976, 13, 303–349. doi:10.1016/S0079-6468(08)70141-3 |
| 49. | Hacksell, U.; Daves, G. D., Jr. Prog. Med. Chem. 1985, 22, 1–65. doi:10.1016/S0079-6468(08)70228-5 |
| 54. | Lim, M. I.; Ren, W. Y.; Otter, B. A.; Klein, R. S. J. Org. Chem. 1983, 48, 780–788. doi:10.1021/jo00154a005 |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 67. | Lim, M.-I.; Klein, R. S. Tetrahedron Lett. 1981, 22, 25–28. doi:10.1016/0040-4039(81)80031-7 |
| 68. | Ren, W. Y.; Lim, M. I.; Otter, B. A.; Klein, R. S. J. Org. Chem. 1982, 47, 4633–4637. doi:10.1021/jo00145a005 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 32. | Ho, M.-C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P. C.; Evans, G. B.; Clinch, K.; Almo, S. C.; Schramm, V. L. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 4805–4812. doi:10.1073/pnas.0913439107 |
| 63. | Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; Doerffler, E.; Byun, D.; Wang, T.; Babusis, D.; Carey, A. C.; German, P.; Sauer, D.; Zhong, W.; Rossi, S.; Fenaux, M.; McHutchison, J. G.; Perry, J.; Feng, J.; Ray, A. S.; Kim, C. U. J. Med. Chem. 2014, 57, 1812–1825. doi:10.1021/jm400201a |
| 64. | Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; Larson, N.; Strickley, R.; Wells, J.; Stuthman, K. S.; Van Tongeren, S. A.; Garza, N. L.; Donnelly, G.; Shurtleff, A. C.; Retterer, C. J.; Gharaibeh, D.; Zamani, R.; Kenny, T.; Eaton, B. P.; Grimes, E.; Welch, L. S.; Gomba, L.; Wilhelmsen, C. L.; Nichols, D. K.; Nuss, J. E.; Nagle, E. R.; Kugelman, J. R.; Palacios, G.; Doerffler, E.; Neville, S.; Carra, E.; Clarke, M. O.; Zhang, L.; Lew, W.; Ross, B.; Wang, Q.; Chun, K.; Wolfe, L.; Babusis, D.; Park, Y.; Stray, K. M.; Trancheva, I.; Feng, J. Y.; Barauskas, O.; Xu, Y.; Wong, P.; Braun, M. R.; Flint, M.; McMullan, L. K.; Chen, S.-S.; Fearns, R.; Swaminathan, S.; Mayers, D. L.; Spiropoulou, C. F.; Lee, W. A.; Nichol, S. T.; Cihlar, T.; Bavari, S. Nature 2016, 531, 381–385. doi:10.1038/nature17180 |
| 65. | Siegel, D.; Hui, H. C.; Doerffler, E.; Clarke, M. O.; Chun, K.; Zhang, L.; Neville, S.; Carra, E.; Lew, W.; Ross, B.; Wang, Q.; Wolfe, L.; Jordan, R.; Soloveva, V.; Knox, J.; Perry, J.; Perron, M.; Stray, K. M.; Barauskas, O.; Feng, J. Y.; Xu, Y.; Lee, G.; Rheingold, A. L.; Ray, A. S.; Bannister, R.; Strickley, R.; Swaminathan, S.; Lee, W. A.; Bavari, S.; Cihlar, T.; Lo, M. K.; Warren, T. K.; Mackman, R. L. J. Med. Chem. 2017, 60, 1648–1661. doi:10.1021/acs.jmedchem.6b01594 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
| 48. | Daves, G. D., Jr.; Cheng, C. C. Prog. Med. Chem. 1976, 13, 303–349. doi:10.1016/S0079-6468(08)70141-3 |
| 49. | Hacksell, U.; Daves, G. D., Jr. Prog. Med. Chem. 1985, 22, 1–65. doi:10.1016/S0079-6468(08)70228-5 |
| 62. | Štambaský, J.; Hocek, M.; Kočovský, P. Chem. Rev. 2009, 109, 6729–6764. doi:10.1021/cr9002165 |
| 74. | Wang, G.; Wan, J.; Hu, Y.; Wu, X.; Prhavc, M.; Dyatkina, N.; Rajwanshi, V. K.; Smith, D. B.; Jekle, A.; Kinkade, A.; Symons, J. A.; Jin, Z.; Deval, J.; Zhang, Q.; Tam, Y.; Chanda, S.; Blatt, L.; Beigelman, L. J. Med. Chem. 2016, 59, 4611–4624. doi:10.1021/acs.jmedchem.5b01933 |
© 2018 Temburnikar and Seley-Radtke; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (https://www.beilstein-journals.org/bjoc)