Abstract
The chemical similarity of antibacterial cyclic peptides and peptidomimetics was studied in order to identify new promising cyclic scaffolds. A large descriptor space coupled with cluster analysis was employed to digitize known antibacterial structures and to gauge the potential of new peptidomimetic macrocycles, which were conveniently synthesized by acylbenzotriazole methodology. Some of the synthesized compounds were tested against an array of microorganisms and showed antibacterial activity against Bordetella bronchistepica, Micrococcus luteus, and Salmonella typhimurium.
Graphical Abstract
Introduction
Diverse cyclic peptides, both natural and synthesized, are potent antibiotics [1-3]. Gramicidin S, vancomycine, bacitracin, polymixin B, colistin, valinomycin, actinomycin, and many more, have been tested and used clinically as antimicrobial and antifungal agents: It is believed that cyclic peptides are more stable to proteolysis due to the lack of free N- and C-termini, as well as reduced conformational freedom. Stability can also be achieved by modifying peptides into “peptidomimetics” that mimic and/or stabilize the secondary structure that modifies associated biological processes, thus affording opportunities for drug design and development.
One strategy to create peptidomimetics couples a small-molecule scaffold with a peptide. Such scaffolds include aromatic rings or heterocycles, which may be positioned in the interior of the peptide chain [4,5] or at the C- [6-8] or N-terminus [9]. Many reports describe the successful use of heterocycles as peptide-bond surrogates or as potential protein-recognition motifs to achieve superior potency in biological assays [10-14].
Pyridines are well-established as important heterocycles in medicinally and biologically active compounds and also as important structural elements. Pyridine scaffolds possess important antiviral [15], anti-inflammatory [16,17], anticonvulsant [18], antibacterial [19], and antitumor pharmacological activities [20,21].
Chiral macrocyclic ligands have found wide application in asymmetric synthesis and enantiomeric recognition [22,23]. Incorporation of amino acids in abiotic anion receptors can lead to systems that mimic the anion coordination properties of anion-binding proteins [24]. Introduction of cysteine subunits into a macrocycle facilitates receptor synthesis and allows control of the relative direction of the two chains attached to the cysteine residue. For example, one of the early cysteine-containing macrocycles was designed to mimic the cation binding ability of valinomycin [24].
Numerous literature approaches to the synthesis of 17-, 18- and 24-membered rings utilize: (i) acyl chlorides, (ii) active esters and (iii) coupling reagents. However, some of the published literature methods involve the utilization of complex procedures, protection/deprotection strategies, harsh reaction conditions, and long reaction times; provide only low yields; and are plagued with difficulties associated with product purification [25-29]. According to the literature [30,31], most antimicrobial cyclic peptides are active in their cationic form. Cationic peptides are considered advantageous, because they adhere better to the outer anionic parts of phospholipid membranes.
Although anionic peptides are less common in clinical practice, they are gaining momentum, because natural anionic peptides are a part of the innate immune system of living organisms (located in human, ovine, and bovine lungs) and also serve as immunomodulators [32]. The activity of anionic peptides can be enhanced when associated with zinc cations and/or surfactants [30-32]. Anionic peptides bind with lysozyme, an important cationic hydrolytic enzyme. It is believed that the lysozyme opens the cell wall, “allowing a small anionic peptide to penetrate”. Topical administration of such anionic peptides is possible, for example in the form of nasal or throat sprays but, at least one anionic peptide, daptomycin, is administered intravenously.
The abundance of natural and synthetic antimicrobial peptides and peptidomimetics of varying structure and activity reported raises the question of how their similarity or dissimilarity can be quantified and how synthetic efforts can be focused towards novel active compounds. It would be beneficial to be able to draw practical conclusions (even if approximate) based solely on the chemical structures of existing agents. In the case of cyclic peptides, the complex cyclomatic structure, involving smaller rings and heteroatoms, as well as a limited conformational flexibility, makes it difficult to classify or compare such compounds by their ring sizes. Fortunately, chemical informatics provides formalized procedures for comparing chemical compounds based on composition and structure. While the principle of molecular similarity is quite simple, that is, “similar compounds have similar properties”, the unambiguous and automated implementation of this principle needed to be backed with a solid mathematical background; this has been accomplished and has made it possible to develop efficient similarity measures [33]. The simplest similarity measure of two molecules would be the Euclidian distance between them in a descriptor space, which is an abstract n-dimensional space spanned by n molecular descriptors, or parameters. More elaborate measures have been developed, such as the Tanimoto coefficient or the Tversky index [34]. Although a well-established concept, molecular similarity has previously been used almost entirely in the design of small-molecule drugs, with very few applications in other chemical disciplines. To the best of our knowledge, despite their promise of consistent analysis and comparison, the principles of molecular similarity have never been used in the design of peptides.
The aim of the present work is thus three-fold: (i) To identify promising peptidomimetic scaffolds by using molecular similarity; (ii) to design a range of cyclic pyridine-containing peptidomimetics capable of facile synthesis; and (iii) to synthesize and biotest promising candidates. A synthetic route seems highly relevant, since N-acylbenzotriazoles have been reported elsewhere [35-37] to be stable, easy-to-handle acylating agents and have found numerous applications for advantageous N-, O-, C- and S-acylations.
Results and Discussion
Similarity analysis
Our literature search identified the thirty three cyclic peptides and peptidomimetic structures given in Table 1. All the minimum inhibition constants (MIC) are pertinent to Staphylococcus aureus. These include (i) one 12-membered ring cyclic tetrapeptide, (ii) two 14-membered ring peptidomimetics each with two peptide links, (iii) two 18-membered ring peptidomimetics each with five peptide links, and (iv) twenty seven 16-membered ring pentapeptides. Table 1 demonstrates that compounds of group (iv) show the best antibacterial activity, with compounds 24 and 27 being the most active. The side-chain substitution clearly controls the antibacterial activity, as the cyclic scaffold is identical. The 18-membered ring scaffold 37, accessible through the benzotriazole route, provides an excellent opportunity for introducing substituents economically, through the proper choice of the dicarboxylic starting materials. While scaffold 37 and the 16-membered ring scaffolds described in Table 1 seem to be visually different, we undertook analysis of their proximity in a multidimensional descriptor space to afford a more rigorous comparison. Such analysis takes into account not only structural similarity features, but also electronic, charge-distribution, and hydrogen-bond characteristics. We generated such a multidimensional descriptor space for all structures in Table 1 as well as for structures 37a–c, using CODESSA-Pro software [38]. Clustering of the structures in such a multidimensional space is the natural choice for similarity analysis.
Table 1: Structures of cyclic peptides with their MIC values.
| Structure | Cluster number | Observed MIC (μg/mL) | Ref. |
|---|---|---|---|
|
1 |
3 | 8.0 | [39] |
|
2 |
2 | 16 | [39] |
|
3 |
3 | -- | [40] |
|
4 |
3 | 3.12 | [41] |
|
5 |
3 | 3.12 | [42] |
|
6 |
2 | 0.50 | [43,44] |
|
7 |
3 | 1.5 | [44] |
|
8 |
1 | 12.5 | [45] |
|
9 |
2 | 50 | [45] |
|
10 |
1 | 12.5 | [45] |
|
11 |
2 | 1.56 | [45] |
|
12 |
1 | 1.56 | [45] |
|
13 |
2 | 6.25 | [46] |
|
14 |
2 | 1.56 | [46] |
|
15 |
2 | 3.12 | [47] |
|
16 |
2 | 12.5 | [47] |
|
17 |
2 | 12.5 | [47] |
|
18 |
2 | 12.5 | [47] |
|
19 |
2 | 12.5 | [47] |
|
20 |
1 | 25.0 | [47] |
|
21 |
2 | 25.0 | [47] |
|
22 |
2 | 12.5 | [47] |
|
23 |
2 | 12.5 | [47] |
|
24 |
2 | 0.78 | [48] |
|
25 |
2 | 6.25 | [48] |
|
26 |
1 | 1.56 | [48] |
|
27 |
2 | 0.780 | [48] |
|
28 |
2 | 3.12 | [48] |
|
29 |
2 | 1.56 | [48] |
|
30 |
2 | 1.56 | [48] |
|
31 |
2 | 3.12 | [48] |
|
32 |
2 | 3.12 | [48] |
|
33 |
2 | 3.12 | [48] |
Clustering
Clustering is a widely used technique that has found application in the selection of compounds for screening, analysis of substructure search output, and the prediction of molecular properties [49]. The “k-means method” implemented in Statistica software is effective in clustering results for many practical applications, and was used to classify the current data set. All types of descriptors are calculated by using the CODESSA Pro program and the descriptor matrix is generated, which served as an input for the cluster-analysis module in STATISTICA software. The use of descriptors in this study can give differing clustering patterns [50]. The first step in the structure clustering is the generation of a matrix containing similarity values of descriptors for all pairs of compounds. Further, the matrix is converted to a hierarchical clustering tree by using single-linkage amalgamation and Euclidian distance measure. The single-linkage approach assigns sequential cluster labels to nearest-neighbor clusters. Then a series of clustering experiments was carried out with different cutoffs on the descriptor dependence, to clearly establish whether parameters produce realistic improvements in the quality of the results or not [51].
Analysis of the membership functions shows that the clusters are well-defined with each compound typically having one membership function, as shown in Figure 1 and Table 1. The cluster analysis results demonstrate that most of the studied compounds are clustered clusters. The peptidomimetics 37a–c proposed for synthesis are located in cluster 1. Cluster 1 mainly consists of both cyclotetra- and cyclopentapeptides that are similar in ring size to 37a–c; this cluster also includes those cyclic peptides that are highly potent as antibacterial agents (smallest MIC values). Clusters 1 and 2 contain structurally more diverse peptides. Thus, the majority of 12-membered ring-cyclic compounds were assigned to cluster 1, while the more branched structures with longer side chains were found in cluster 2. These assignments are chemically meaningful and thus bear witness to the validity of the clusterization method used.
Figure 1: Molecular-descriptor-based cluster analysis; single-linkage Euclidean distances. Clustering of compounds in the descriptor space; a two-dimensional representation of chemical space being partitioned into clusters of similar compounds based on descriptors using a top-down (hierarchical) clustering method. Structures for compounds 37a–37c are shown in Scheme 2. (Axis X denotes compounds and axis Y denotes linkage distance); the red oval, blue rectangle, and green oval represent cluster 1, cluster 2, and cluster 3, respectively.
Figure 1: Molecular-descriptor-based cluster analysis; single-linkage Euclidean distances. Clustering of comp...
The proximity of the cyclic peptidomimetics under study (37a–c) and the existing antibacterial cyclic peptides in such a rich descriptor space, in which all major structural, electronic, and intermolecular-interaction features are taken into account, attests to the feasibility of using the 17- and 18-membered ring scaffolds 37 as a main building block for designing novel antibacterial agents. This, along with the facile and economic synthetic route, renders scaffold 37 as a promising platform for further rational drug design.
Synthesis
Treatment of dicarboxylic acids 34a–c by a standard method [36] using thionyl chloride and 1H-benzotriazole gave the corresponding benzotriazole derivatives in 37–54% yield (Scheme 1, see Supporting Information File 1 for experimental details).
Scheme 1: Preparation of dicarboxylic benzotriazole derivatives.
Scheme 1: Preparation of dicarboxylic benzotriazole derivatives.
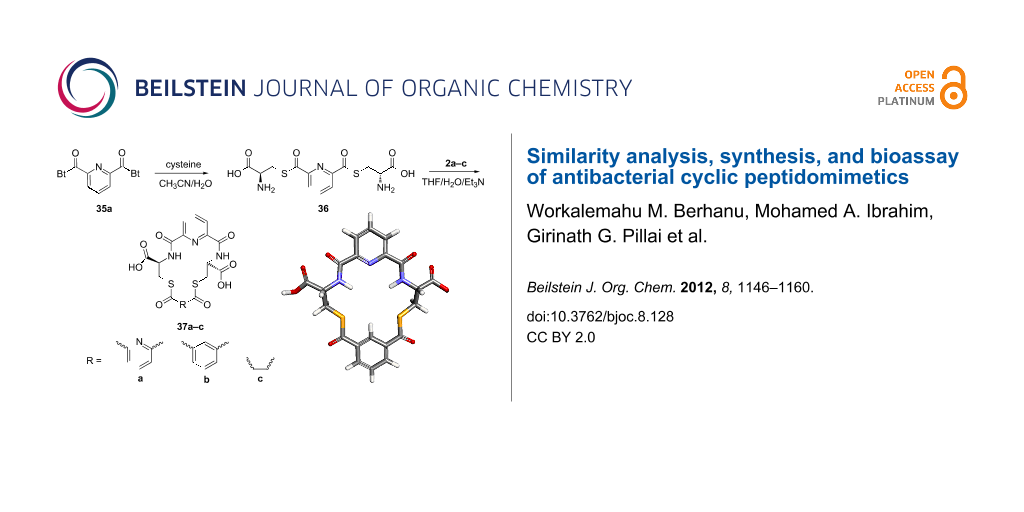
The methodology used for the regioselective syntheses of S- and N-acylcysteines was developed recently in our group by using N-acylbenzotriazoles under mild reaction conditions [37]. Utilizing this methodology, 35a was coupled with two equiv of free cysteine in aqueous acetonitrile at 20 °C over 12 h to give bis(S-acylcysteine) 36 in 64% yield. Compound 36 was then treated with 1 equiv of 35a–c to synthesize cyclic enantiopure peptidomimetic products 37a–c in 81–82% yield (Scheme 2, see Supporting Information File 1 for experimental details).
Scheme 2: Preparation of pyridine-based cysteine-containing macrocycles.
Scheme 2: Preparation of pyridine-based cysteine-containing macrocycles.
In a further application of this synthetic approach 35 was coupled with 2 equiv of L-Phe-OH in the presence of TEA in aqueous acetonitrile at room temperature over 3 h giving the bis N-acylated compound 38 in 88% yield. Compound 38 was converted to the corresponding benzotriazole derivative 39 and coupled with bis(S-acylcysteine) 36 forming the pyridine–cysteine-containing macrocycle 40 in 70% yield (Scheme 3, see Supporting Information File 1 for experimental details).
Scheme 3: Preparation of pyridine–cysteine-containing macrocycle 39.
Scheme 3: Preparation of pyridine–cysteine-containing macrocycle 39.
Bioassay
Screening for antibacterial activity was performed for two cyclic peptidomimetics belonging to scaffold 37, namely 37a and 37b, with the pyridine- and phenyl-linking fragments, respectively. The array of microbial strains assayed in this study included six pathogens listed in Table 2 (see Supporting Information File 1, for antibacterial testing protocol).
Table 2: Antibacterial activity in vitro.
| Microbial straina | Inhibition zone (mm) at 200 µg/mL | MICb (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 37a | 37b | Roxc | Cfxd | 37a | 37b | Roxc | Cfxd | ||
|
Staphylococcus aureus
(ATCC 6538) |
0.0 | 0.0 | 30 | 40 | – | – | 12.5 | 6.25 | |
|
Escherichia coli
(ATCC 8739) |
0.0 | 0.0 | 14.6 | 22.9 | – | – | 100 | 50 | |
|
Bordetella bronchiseptica
(ATCC 4617) |
0.0 | 6.0 | 8 | 33 | – | 150 | 150 | 12.5 | |
|
Micrococcus luteus
(ATCC 10240) |
0.0 | 6.0 | 30 | 40 | – | 150 | 12.5 | 12.5 | |
|
Salmonella typhimurium
(ATCC 14028) |
0.0 | 6.0 | 12 | 40 | – | 150 | 100 | 6.25 | |
|
Enterobacter aerogenes
(ATCC 13048) |
0.0 | 0.0 | 15 | 34 | – | – | 100 | 12.5 | |
aBordetella bronchistepica: Gram negative, relevant to veterinary science; Micrococcus luteus: Gram positive, not dangerous for humans; Salmonella typhimurium: Gram negative, relevant to veterinary science. bMIC = minimum inhibitory concentration. cRox = roxithromycin. dCfx = cefixime.
The in vitro data for 37a and 37b are given in Table 2. Reference antibiotics roxithromycin and cefixime were used as positive controls. It is seen that peptidomimetic 37b exhibits moderate activity against Bordetella bronchiseptica, Micrococcus luteus, and Salmonella typhimurium. For a “bare” scaffold with no tailor-made substitution, this should be considered as an encouraging result and an indication of the possibility of more interesting results if the scaffold is furnished with appropriate functions.
Peptidomimetic 37a showed no activity in this test, which can be explained by the formation of a zwitter-ionic structure due to protonation of the pyridine moiety by free carboxylic groups. It is reasonable to assume that such a zwitter-ionic structure alters the charge distribution and also eliminates hydrogen bonding in which the pyridine nitrogen atom acts as a H-bond acceptor. These factors increase hydrophobicity and thus deteriorate bioavailability, as reflected in the absence of activity of 37a. Absence of data in Table 2 means that the sample is not active at the highest concentration tested.
Conclusion
Two new cyclic peptidomimetic scaffolds were identified by using similarity-based rational design. Their chemical similarity with existing antibacterial cyclic peptidomimetics was established in a huge descriptor space generated by Codessa-Pro software. At least one compound (37b) demonstrated a moderate antibacterial activity against three bacterial strains, which is a fairly promising result for a “bare” cyclic scaffold with no intentional functionalization. Given the successful scaffold identified, the next step will be to furnish it with appropriate functional groups and substituents by using rational design principles.
Experimental
Methodology
Molecular similarity
The potential of cyclic peptidomimetics, widely acknowledged as significant, arises because they are less prone to hydrolysis in vivo and can be synthesized rather inexpensively. The synthetic methodology that has been developed in our lab affords facile assembles of cyclic structures from dicarboxylic building blocks by using a benzotriazole methodology. Seventeen and eighteen-membered sulfur-containing rings can be synthesized readily if cysteine is used as the coupling agent. Such facile access to cyclic peptidomimetics is appropriate for the design of compounds expected to be biologically active as antibacterial agents. Exploring the molecular similarity of existing antibacterial cyclic peptides using the Tanimoto method in the Instant JChem software [52], and the peptidomimetics easily accessible through the benzotriazole route, should give structural insights and design criteria. Guidance for such a similarity-based design should be available by analysis of a reasonably sized dataset of existing analogues. As abundant data can be found in the literature for Staphylococcus aureus, this was chosen as a reference for antibacterial activity. SciFinder Scholar was used to retrieve the structures of 33 cyclic peptides with reported antibacterial activity (Table 1). The minimum growth inhibition constant (MIC) was used as the measure of antibacterial activity, and the data collected were converted where necessary to μg/mL. Three structures newly synthesized in our lab were also added to the general dataset (Table 1). All these structures were drawn with the Marvin Beans Suite program [52] and preoptimized using the molecular mechanics utility (MM2) [53] in Chem3D Ultra 12.0 software [54]. Final geometry optimization of the compounds was carried out by using the semiempirical quantum-mechanical AM1 parameterization [55].
Molecular descriptors
The optimized geometries of the compounds were loaded into CODESSA Pro software [38]. Overall, more than 800 theoretical descriptors were calculated including constitutional, geometrical, topological, electrostatic, quantum-chemical, and thermodynamic molecular descriptors [56,57].
Cluster analysis
Cluster analysis of the descriptor hyperspace was performed by using STATISTICA version 6 software [58], with Euclidean distance and other metrics used as similarity measures. Cluster analysis guided by the experimental MIC available for some compounds should help reveal structural features affording antibacterial activity and to identify hotspots in the descriptor hyperspace. Combinations of structural features in such hotspots can be used as guidelines for the rational design of cyclic peptide structures to achieve desirable levels of antibacterial activity.
Supporting Information
| Supporting Information File 1: Experimental details, characterization data of synthesized compounds and antibacterial testing protocol. | ||
| Format: PDF | Size: 170.9 KB | Download |
Acknowledgements
We thank the University of Florida, the Kenan Foundation and King Abdulaziz University, Jeddah, Saudi Arabia for financial support. We are grateful to Mr. Alexander Drijver, ChemAxon Kft. Hungary for a free academic license. The authors are grateful to Dr. C. D. Hall and Ms. Galyna Vakulenko for useful discussions and help.
References
-
Powers, J.-P. S.; Hancock, R. E. W. Peptides 2003, 24, 1681–1691. doi:10.1016/j.peptides.2003.08.023
Return to citation in text: [1] -
Dartois, V.; Sanchez-Quesada, J.; Cabezas, E.; Chi, E.; Dubbelde, C.; Dunn, C.; Granja, J.; Gritzen, C.; Weinberger, D.; Ghadiri, M. R.; Parr, T. R., Jr. Antimicrob. Agents Chemother. 2005, 49, 3302–3310. doi:10.1128/AAC.49.8.3302-3310.2005
Return to citation in text: [1] -
Epand, R. M.; Vogel, H. J. Biochim. Biophys. Acta 1999, 1462, 11–28. doi:10.1016/S0005-2736(99)00198-4
Return to citation in text: [1] -
Singh, A.; Wilczynski, A.; Holder, J. R.; Witek, R. M.; Dirain, M. L.; Xiang, Z.; Edison, A. S.; Haskell-Luevano, C. J. Med. Chem. 2011, 54, 1379–1390. doi:10.1021/jm101425m
Return to citation in text: [1] -
Bach, A.; Strømgaard, K. Synthesis 2011, 807–815. doi:10.1055/s-0030-1258425
Return to citation in text: [1] -
Biron, E.; Chatterjee, J.; Kessler, H. Org. Lett. 2006, 8, 2417–2420. doi:10.1021/ol0607645
Return to citation in text: [1] -
Tornøe, C. W.; Sanderson, S. J.; Mottram, J. C.; Coombs, G. H.; Meldal, M. J. Comb. Chem. 2004, 6, 312–324. doi:10.1021/cc020085v
Return to citation in text: [1] -
Saitton, S.; Kihlberg, J.; Luthman, K. Tetrahedron 2004, 60, 6113–6120. doi:10.1016/j.tet.2004.05.048
Return to citation in text: [1] -
McComsey, D. F.; Hawkins, M. J.; Andrade-Gordon, P.; Addo, M. F.; Oksenberg, D.; Maryanoff, B. E. Bioorg. Med. Chem. Lett. 1999, 9, 1423–1428. doi:10.1016/S0960-894X(99)00197-3
Return to citation in text: [1] -
Pedersen, D. S.; Abell, A. Eur. J. Org. Chem. 2011, 2399–2411. doi:10.1002/ejoc.201100157
Return to citation in text: [1] -
Appendino, G.; Bacchiega, S.; Minassi, A.; Cascio, M. G.; De Petrocellis, L.; Di Marzo, V. D. Angew. Chem., Int. Ed. 2007, 46, 9312–9315. doi:10.1002/anie.200703590
Return to citation in text: [1] -
Tam, A.; Arnold, U.; Soellner, M. B.; Raines, R. T. J. Am. Chem. Soc. 2007, 129, 12670–12671. doi:10.1021/ja075865s
Return to citation in text: [1] -
Brik, A.; Yang, Y.-Y.; Ficht, S.; Wang, C.-H. J. Am. Chem. Soc. 2006, 128, 5626–5627. doi:10.1021/ja061165w
Return to citation in text: [1] -
Borg, S.; Vollinga, R. C.; Labarre, M.; Payza, K.; Terenius, L.; Luthman, K. J. Med. Chem. 1999, 42, 4331–4342. doi:10.1021/jm990197+
Return to citation in text: [1] -
Balzarini, J.; Stevens, M.; De Clercq, E.; Schols, D.; Pannecouque, C. J. Antimicrob. Chemother. 2005, 55, 135–138. doi:10.1093/jac/dkh530
Return to citation in text: [1] -
Haviv, F.; DeNet, R. W.; Michaels, R. J.; Ratajczyk, J. D.; Carter, G. W.; Young, P. R. J. Med. Chem. 1983, 26, 218–222. doi:10.1021/jm00356a018
Return to citation in text: [1] -
Thirumurugan, P.; Mahalaxmi, S.; Perumal, P. T. J. Chem. Sci. 2010, 122, 819–832. doi:10.1007/s12039-010-0070-3
Return to citation in text: [1] -
Phillips, O. A.; Knaus, E. E. Drug Des. Delivery 1991, 4, 279–286.
Return to citation in text: [1] -
Reck, F.; Zhou, F.; Eyermann, C. J.; Kem, G.; Carcanague, D.; Ioannidis, G.; Illingworth, R.; Poon, G.; Gravestock, M. B. J. Med. Chem. 2007, 50, 4868–4881. doi:10.1021/jm070428+
Return to citation in text: [1] -
Nassar, E. J. Am. Sci. 2010, 6 (8), 463–471.
Return to citation in text: [1] -
Liu, M. C.; Lin, T. C.; Sartorelli, A. C. J. Med. Chem. 1992, 35, 3672–3677. doi:10.1021/jm00098a012
Return to citation in text: [1] -
Kim, G.-J.; Park, D.-W.; Tak, Y.-S. Catal. Lett. 2000, 65, 127–133. doi:10.1023/A:1019040532103
Return to citation in text: [1] -
Li, Z.; Jablonski, C. Chem. Commun. 1999, 1531–1532. doi:10.1039/a900018f
Return to citation in text: [1] -
Kubik, S. Chem. Soc. Rev. 2009, 38, 585–605. doi:10.1039/b810531f
Return to citation in text: [1] [2] -
Ranganathan, D.; Thomas, A.; Haridas, V.; Kurur, S.; Madhusudanan, K. P.; Roy, R.; Kunwar, A. C.; Sarma, A. V. S.; Vairamani, M.; Sarma, K. D. J. Org. Chem. 1999, 64, 3620–3629. doi:10.1021/jo982472q
Return to citation in text: [1] -
Ranganathan, D.; Haridas, V.; Gilardi, R.; Karle, I. L. J. Am. Chem. Soc. 1998, 120, 10793–10800. doi:10.1021/ja982244d
Return to citation in text: [1] -
Pintér, Á.; Haberhauer, G. Tetrahedron 2009, 65, 2217–2225. doi:10.1016/j.tet.2009.01.047
Return to citation in text: [1] -
Montero, A.; Albericio, F.; Royo, M.; Herradon, B. Eur. J. Org. Chem. 2007, 1301–1308. doi:10.1002/ejoc.200600833
Return to citation in text: [1] -
Wang, X.; Wang, Q.; Huang, X.; Wang, T.; Yu, X. ARKIVOC 2006, xi, 1–7.
Return to citation in text: [1] -
Hancock, R. E. W. Lancet Infect. Dis. 2001, 1, 156–164. doi:10.1016/S1473-3099(01)00092-5
Return to citation in text: [1] [2] -
Wu, M. H.; Hancock, R. E. W. Antimicrob. Agents Chemother. 1999, 43, 1274–1276.
Return to citation in text: [1] [2] -
Fales-Williams, A. J.; Gallup, J. M.; Ramirez-Romero, R.; Brogden, K. A.; Ackermann, M. R. Clin. Diagn. Lab. Immunol. 2002, 9, 28–32. doi:10.1128/CDLI.9.1.28-32.2002
Return to citation in text: [1] [2] -
Alastair, A. R. Generalized Method of Moments; Oxford University Press: Oxford, U.K., 2005.
Return to citation in text: [1] -
Bender, A.; Glen, R. C. Org. Biomol. Chem. 2004, 2, 3204–3218. doi:10.1039/b409813g
Return to citation in text: [1] -
Katritzky, A. R.; Khelashvili, L.; Mohapatra, P. P.; Steel, P. J. Synthesis 2007, 3673–3677. doi:10.1055/s-2007-990889
Return to citation in text: [1] -
Katritzky, A. R.; Angrish, P.; Todadze, E. Synlett 2009, 2392–2411. doi:10.1055/s-0029-1217827
Return to citation in text: [1] [2] -
Katritzky, A. R.; Tala, S. R.; Abo-Dya, N. E.; Gyanda, K.; El-Gendy, B. E.-D. M.; Abdel-Samii, Z. K.; Steel, P. J. J. Org. Chem. 2009, 74, 7165–7167. doi:10.1021/jo900853s
Return to citation in text: [1] [2] -
CODESSA Pro; University of Florida: Gainesville, FL, USA, 2001.
Return to citation in text: [1] [2] -
von Nussbaum, F. V.; Anlauf, S.; Freiberg, C.; Benet-Buchholz, J.; Schamberger, J.; Henkel, T.; Schiffer, G.; Häbich, D. ChemMedChem 2008, 3, 619–626. doi:10.1002/cmdc.200700297
Return to citation in text: [1] [2] -
Morel, A. F.; Maldaner, G.; Ilha, V.; Missau, F.; da Silva, U. F.; Dalcol, I. I. Phytochemistry 2005, 66, 2571–2576. doi:10.1016/j.phytochem.2005.08.016
Return to citation in text: [1] -
Morel, A. F.; Araujo, C. A.; da Silva, U. F.; Hoelzel, S. C. S. M.; Záchia, R.; Bastos, N. R. Phytochemistry 2002, 61, 561–566. doi:10.1016/S0031-9422(02)00287-X
Return to citation in text: [1] -
Giacomelli, S. R.; Maldaner, G.; Gonzaga, W. A.; Garcia, C. M.; da Silva, U. F.; Dalcol, I. I.; Morel, A. F. Phytochemistry 2004, 65, 933–937. doi:10.1016/j.phytochem.2004.02.006
Return to citation in text: [1] -
Socha, A. M.; Tan, N. Y.; LaPlante, K. L.; Sello, J. K. Bioorg. Med. Chem. 2010, 18, 7193–7202. doi:10.1016/j.bmc.2010.08.032
Return to citation in text: [1] -
von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Angew. Chem., Int. Ed. 2006, 45, 5072–5129. doi:10.1002/anie.200600350
Return to citation in text: [1] [2] -
Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1
Return to citation in text: [1] [2] [3] [4] [5] -
Norcia, L. J. L.; Silvia, A. M.; Dirlam, J. P.; Schnur, R. C.; Bergeron, J. M.; Retsema, J. A.; Hayashi, S. F. J. Antibiot. 1999, 52, 1007–1016. doi:10.7164/antibiotics.52.1007
Return to citation in text: [1] [2] -
Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] -
Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] -
Holliday, J. D.; Rodgers, S. L.; Willett, P.; Chen, M. Y.; Mahfouf, M.; Lawson, K. J. Chem. Inf. Comput. Sci. 2004, 44, 894–902. doi:10.1021/ci0342674
Return to citation in text: [1] -
Stahl, M.; Mauser, H.; Tsui, M.; Taylor, N. R. J. Med. Chem. 2005, 48, 4358–4366. doi:10.1021/jm040213p
Return to citation in text: [1] -
Adamson, G. W.; Bawden, D. J. Chem. Inf. Comput. Sci. 1981, 21, 204–209. doi:10.1021/ci00032a005
Return to citation in text: [1] -
Instant JChem 5.5.0; Marvin 5.6.0; ChemAxon Kft.: Hungary, 2011.
Return to citation in text: [1] [2] -
Allinger, N. L. J. Am. Chem. Soc. 1977, 99, 8127–8134. doi:10.1021/ja00467a001
Return to citation in text: [1] -
ChemBioDraw Ultra, Version 12.0.2; Cambridgesoft: Cambridge, MA, USA, 2010.
Return to citation in text: [1] -
Dewar, M. J. S.; Zoebisch, E. G.; Healy, E. F.; Stewart, J. J. P. J. Am. Chem. Soc. 1985, 107, 3902–3909. doi:10.1021/ja00299a024
Return to citation in text: [1] -
Karelson, M. Molecular Descriptors in QSAR/QSPR; Wiley Interscience: New York, 2000.
Return to citation in text: [1] -
Katritzky, A. R.; Kuanar, M.; Slavov, S.; Hall, C. D.; Karelson, M.; Kahn, I.; Dobchev, D. A. Chem. Rev. 2010, 110, 5714–5789. doi:10.1021/cr900238d
Return to citation in text: [1] -
STATISTICA, Version 6; StatSoft, Inc.: Tulsa, OK, USA, 2001.
Return to citation in text: [1]
| 44. | von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Angew. Chem., Int. Ed. 2006, 45, 5072–5129. doi:10.1002/anie.200600350 |
| 45. | Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1 |
| 45. | Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1 |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 46. | Norcia, L. J. L.; Silvia, A. M.; Dirlam, J. P.; Schnur, R. C.; Bergeron, J. M.; Retsema, J. A.; Hayashi, S. F. J. Antibiot. 1999, 52, 1007–1016. doi:10.7164/antibiotics.52.1007 |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 45. | Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1 |
| 46. | Norcia, L. J. L.; Silvia, A. M.; Dirlam, J. P.; Schnur, R. C.; Bergeron, J. M.; Retsema, J. A.; Hayashi, S. F. J. Antibiot. 1999, 52, 1007–1016. doi:10.7164/antibiotics.52.1007 |
| 45. | Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1 |
| 45. | Linde, R. G., II; Birsner, N. C.; Chandrasekaran, R. Y.; Clancy, J.; Howe, R. J.; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Petitpas, J. W.; Rainville, J. P.; Su, W.-G.; Vu, C. B.; Whipple, D. A. Bioorg. Med. Chem. Lett. 1997, 7, 1149–1152. doi:10.1016/S0960-894X(97)00188-1 |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 47. | Lyssikatos, J. P.; Chang, S.-P.; Clancy, J.; Dirlam, J. P.; Finegan, S. M.; Girard, A. E.; Hayashi, S. F.; Larson, D. P.; Lee, A. S.; Linde, R. G., II; MacLelland, C. P.; Petipas, J. W.; Seibel, S. B.; Vu, C. B. Bioorg. Med. Chem. Lett. 1997, 7, 1145–1148. doi:10.1016/S0960-894X(97)00187-X |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 1. | Powers, J.-P. S.; Hancock, R. E. W. Peptides 2003, 24, 1681–1691. doi:10.1016/j.peptides.2003.08.023 |
| 2. | Dartois, V.; Sanchez-Quesada, J.; Cabezas, E.; Chi, E.; Dubbelde, C.; Dunn, C.; Granja, J.; Gritzen, C.; Weinberger, D.; Ghadiri, M. R.; Parr, T. R., Jr. Antimicrob. Agents Chemother. 2005, 49, 3302–3310. doi:10.1128/AAC.49.8.3302-3310.2005 |
| 3. | Epand, R. M.; Vogel, H. J. Biochim. Biophys. Acta 1999, 1462, 11–28. doi:10.1016/S0005-2736(99)00198-4 |
| 10. | Pedersen, D. S.; Abell, A. Eur. J. Org. Chem. 2011, 2399–2411. doi:10.1002/ejoc.201100157 |
| 11. | Appendino, G.; Bacchiega, S.; Minassi, A.; Cascio, M. G.; De Petrocellis, L.; Di Marzo, V. D. Angew. Chem., Int. Ed. 2007, 46, 9312–9315. doi:10.1002/anie.200703590 |
| 12. | Tam, A.; Arnold, U.; Soellner, M. B.; Raines, R. T. J. Am. Chem. Soc. 2007, 129, 12670–12671. doi:10.1021/ja075865s |
| 13. | Brik, A.; Yang, Y.-Y.; Ficht, S.; Wang, C.-H. J. Am. Chem. Soc. 2006, 128, 5626–5627. doi:10.1021/ja061165w |
| 14. | Borg, S.; Vollinga, R. C.; Labarre, M.; Payza, K.; Terenius, L.; Luthman, K. J. Med. Chem. 1999, 42, 4331–4342. doi:10.1021/jm990197+ |
| 30. | Hancock, R. E. W. Lancet Infect. Dis. 2001, 1, 156–164. doi:10.1016/S1473-3099(01)00092-5 |
| 31. | Wu, M. H.; Hancock, R. E. W. Antimicrob. Agents Chemother. 1999, 43, 1274–1276. |
| 37. | Katritzky, A. R.; Tala, S. R.; Abo-Dya, N. E.; Gyanda, K.; El-Gendy, B. E.-D. M.; Abdel-Samii, Z. K.; Steel, P. J. J. Org. Chem. 2009, 74, 7165–7167. doi:10.1021/jo900853s |
| 9. | McComsey, D. F.; Hawkins, M. J.; Andrade-Gordon, P.; Addo, M. F.; Oksenberg, D.; Maryanoff, B. E. Bioorg. Med. Chem. Lett. 1999, 9, 1423–1428. doi:10.1016/S0960-894X(99)00197-3 |
| 32. | Fales-Williams, A. J.; Gallup, J. M.; Ramirez-Romero, R.; Brogden, K. A.; Ackermann, M. R. Clin. Diagn. Lab. Immunol. 2002, 9, 28–32. doi:10.1128/CDLI.9.1.28-32.2002 |
| 6. | Biron, E.; Chatterjee, J.; Kessler, H. Org. Lett. 2006, 8, 2417–2420. doi:10.1021/ol0607645 |
| 7. | Tornøe, C. W.; Sanderson, S. J.; Mottram, J. C.; Coombs, G. H.; Meldal, M. J. Comb. Chem. 2004, 6, 312–324. doi:10.1021/cc020085v |
| 8. | Saitton, S.; Kihlberg, J.; Luthman, K. Tetrahedron 2004, 60, 6113–6120. doi:10.1016/j.tet.2004.05.048 |
| 51. | Adamson, G. W.; Bawden, D. J. Chem. Inf. Comput. Sci. 1981, 21, 204–209. doi:10.1021/ci00032a005 |
| 4. | Singh, A.; Wilczynski, A.; Holder, J. R.; Witek, R. M.; Dirain, M. L.; Xiang, Z.; Edison, A. S.; Haskell-Luevano, C. J. Med. Chem. 2011, 54, 1379–1390. doi:10.1021/jm101425m |
| 5. | Bach, A.; Strømgaard, K. Synthesis 2011, 807–815. doi:10.1055/s-0030-1258425 |
| 25. | Ranganathan, D.; Thomas, A.; Haridas, V.; Kurur, S.; Madhusudanan, K. P.; Roy, R.; Kunwar, A. C.; Sarma, A. V. S.; Vairamani, M.; Sarma, K. D. J. Org. Chem. 1999, 64, 3620–3629. doi:10.1021/jo982472q |
| 26. | Ranganathan, D.; Haridas, V.; Gilardi, R.; Karle, I. L. J. Am. Chem. Soc. 1998, 120, 10793–10800. doi:10.1021/ja982244d |
| 27. | Pintér, Á.; Haberhauer, G. Tetrahedron 2009, 65, 2217–2225. doi:10.1016/j.tet.2009.01.047 |
| 28. | Montero, A.; Albericio, F.; Royo, M.; Herradon, B. Eur. J. Org. Chem. 2007, 1301–1308. doi:10.1002/ejoc.200600833 |
| 29. | Wang, X.; Wang, Q.; Huang, X.; Wang, T.; Yu, X. ARKIVOC 2006, xi, 1–7. |
| 36. | Katritzky, A. R.; Angrish, P.; Todadze, E. Synlett 2009, 2392–2411. doi:10.1055/s-0029-1217827 |
| 19. | Reck, F.; Zhou, F.; Eyermann, C. J.; Kem, G.; Carcanague, D.; Ioannidis, G.; Illingworth, R.; Poon, G.; Gravestock, M. B. J. Med. Chem. 2007, 50, 4868–4881. doi:10.1021/jm070428+ |
| 22. | Kim, G.-J.; Park, D.-W.; Tak, Y.-S. Catal. Lett. 2000, 65, 127–133. doi:10.1023/A:1019040532103 |
| 23. | Li, Z.; Jablonski, C. Chem. Commun. 1999, 1531–1532. doi:10.1039/a900018f |
| 49. | Holliday, J. D.; Rodgers, S. L.; Willett, P.; Chen, M. Y.; Mahfouf, M.; Lawson, K. J. Chem. Inf. Comput. Sci. 2004, 44, 894–902. doi:10.1021/ci0342674 |
| 50. | Stahl, M.; Mauser, H.; Tsui, M.; Taylor, N. R. J. Med. Chem. 2005, 48, 4358–4366. doi:10.1021/jm040213p |
| 16. | Haviv, F.; DeNet, R. W.; Michaels, R. J.; Ratajczyk, J. D.; Carter, G. W.; Young, P. R. J. Med. Chem. 1983, 26, 218–222. doi:10.1021/jm00356a018 |
| 17. | Thirumurugan, P.; Mahalaxmi, S.; Perumal, P. T. J. Chem. Sci. 2010, 122, 819–832. doi:10.1007/s12039-010-0070-3 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 15. | Balzarini, J.; Stevens, M.; De Clercq, E.; Schols, D.; Pannecouque, C. J. Antimicrob. Chemother. 2005, 55, 135–138. doi:10.1093/jac/dkh530 |
| 20. | Nassar, E. J. Am. Sci. 2010, 6 (8), 463–471. |
| 21. | Liu, M. C.; Lin, T. C.; Sartorelli, A. C. J. Med. Chem. 1992, 35, 3672–3677. doi:10.1021/jm00098a012 |
| 48. | Dirlam, J. P.; Belton, A. M.; Birsner, N. C.; Brooks, R. R.; Chang, S.-P.; Chandrasekaran, R. Y.; Clancy, J.; Cronin, B. J.; Dirlam, B. P.; Finegan, S. M.; Froshauer, S. A.; Girard, A. E.; Hayashi, S. F.; Howe, R. J.; Kane, J. C.; Kamicker, B. J.; Kaufman, S. A.; Kolosko, N. L.; LeMay, M. A.; Linde, R. G., II; Lyssikatos, J. P.; MacLelland, C. P.; Magee, T. V.; Massa, M. A.; Miller, S. A.; Minich, M. L.; Perry, D. A.; Petitpas, J. W.; Reese, C. P.; Seibel, S. B.; Su, W.-G.; Sweeney, K. T.; Whipple, D. A.; Yang, B. V. Bioorg. Med. Chem. Lett. 1997, 7, 1139–1144. doi:10.1016/S0960-894X(97)00186-8 |
| 34. | Bender, A.; Glen, R. C. Org. Biomol. Chem. 2004, 2, 3204–3218. doi:10.1039/b409813g |
| 30. | Hancock, R. E. W. Lancet Infect. Dis. 2001, 1, 156–164. doi:10.1016/S1473-3099(01)00092-5 |
| 31. | Wu, M. H.; Hancock, R. E. W. Antimicrob. Agents Chemother. 1999, 43, 1274–1276. |
| 32. | Fales-Williams, A. J.; Gallup, J. M.; Ramirez-Romero, R.; Brogden, K. A.; Ackermann, M. R. Clin. Diagn. Lab. Immunol. 2002, 9, 28–32. doi:10.1128/CDLI.9.1.28-32.2002 |
| 33. | Alastair, A. R. Generalized Method of Moments; Oxford University Press: Oxford, U.K., 2005. |
| 53. | Allinger, N. L. J. Am. Chem. Soc. 1977, 99, 8127–8134. doi:10.1021/ja00467a001 |
| 42. | Giacomelli, S. R.; Maldaner, G.; Gonzaga, W. A.; Garcia, C. M.; da Silva, U. F.; Dalcol, I. I.; Morel, A. F. Phytochemistry 2004, 65, 933–937. doi:10.1016/j.phytochem.2004.02.006 |
| 43. | Socha, A. M.; Tan, N. Y.; LaPlante, K. L.; Sello, J. K. Bioorg. Med. Chem. 2010, 18, 7193–7202. doi:10.1016/j.bmc.2010.08.032 |
| 44. | von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Angew. Chem., Int. Ed. 2006, 45, 5072–5129. doi:10.1002/anie.200600350 |
| 40. | Morel, A. F.; Maldaner, G.; Ilha, V.; Missau, F.; da Silva, U. F.; Dalcol, I. I. Phytochemistry 2005, 66, 2571–2576. doi:10.1016/j.phytochem.2005.08.016 |
| 41. | Morel, A. F.; Araujo, C. A.; da Silva, U. F.; Hoelzel, S. C. S. M.; Záchia, R.; Bastos, N. R. Phytochemistry 2002, 61, 561–566. doi:10.1016/S0031-9422(02)00287-X |
| 39. | von Nussbaum, F. V.; Anlauf, S.; Freiberg, C.; Benet-Buchholz, J.; Schamberger, J.; Henkel, T.; Schiffer, G.; Häbich, D. ChemMedChem 2008, 3, 619–626. doi:10.1002/cmdc.200700297 |
| 56. | Karelson, M. Molecular Descriptors in QSAR/QSPR; Wiley Interscience: New York, 2000. |
| 57. | Katritzky, A. R.; Kuanar, M.; Slavov, S.; Hall, C. D.; Karelson, M.; Kahn, I.; Dobchev, D. A. Chem. Rev. 2010, 110, 5714–5789. doi:10.1021/cr900238d |
| 39. | von Nussbaum, F. V.; Anlauf, S.; Freiberg, C.; Benet-Buchholz, J.; Schamberger, J.; Henkel, T.; Schiffer, G.; Häbich, D. ChemMedChem 2008, 3, 619–626. doi:10.1002/cmdc.200700297 |
| 35. | Katritzky, A. R.; Khelashvili, L.; Mohapatra, P. P.; Steel, P. J. Synthesis 2007, 3673–3677. doi:10.1055/s-2007-990889 |
| 36. | Katritzky, A. R.; Angrish, P.; Todadze, E. Synlett 2009, 2392–2411. doi:10.1055/s-0029-1217827 |
| 37. | Katritzky, A. R.; Tala, S. R.; Abo-Dya, N. E.; Gyanda, K.; El-Gendy, B. E.-D. M.; Abdel-Samii, Z. K.; Steel, P. J. J. Org. Chem. 2009, 74, 7165–7167. doi:10.1021/jo900853s |
| 55. | Dewar, M. J. S.; Zoebisch, E. G.; Healy, E. F.; Stewart, J. J. P. J. Am. Chem. Soc. 1985, 107, 3902–3909. doi:10.1021/ja00299a024 |
© 2012 Berhanu et al; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (http://www.beilstein-journals.org/bjoc)