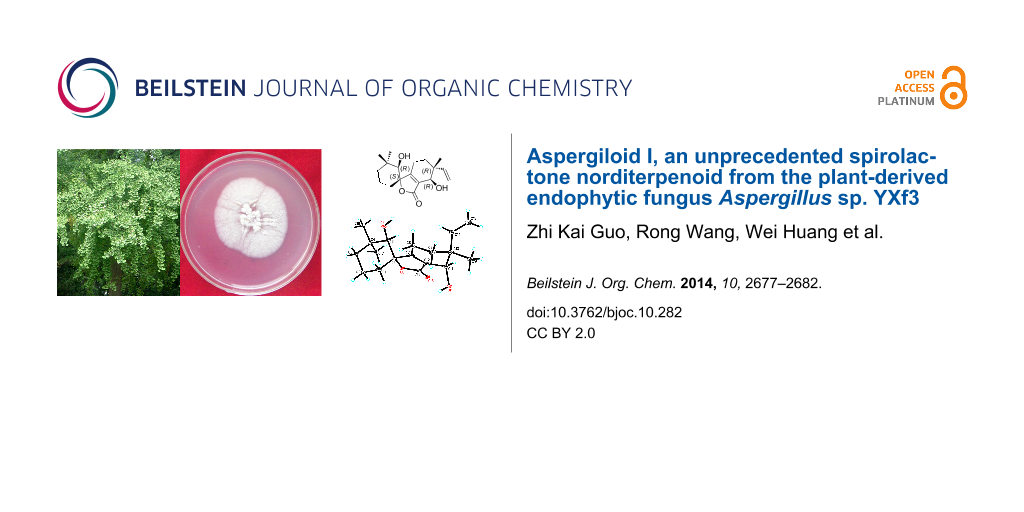
Abstract
An unusual C18 norditerpenoid, aspergiloid I (1), was isolated from the culture broth of Aspergillus sp. YXf3, an endophytic fungus derived from Ginkgo biloba. Its structure was unambiguously established by analysis of HRMS–ESI and spectroscopic data, and the absolute configuration was determined by low-temperature (100 K) single crystal X-ray diffraction with Cu Kα radiation. This compound is structurally characterized by a new carbon skeleton with an unprecedented 6/5/6 tricyclic ring system bearing an α,β-unsaturated spirolactone moiety in ring B, and represents a new subclass of norditerpenoid, the skeleton of which is named aspergilane. The hypothetical biosynthetic pathway for 1 was also proposed. The cytotoxic, antimicrobial, anti-oxidant and enzyme inhibitory activities of 1 were evaluated.
Graphical Abstract
Introduction
Plant-derived fungi, which have drawn considerable attention from natural product chemists, have been proved to be a rich source of bioactive natural compounds [1,2]. Recently, a wide variety of biologically active and structurally unique metabolites were isolated from these types of microorganisms [3-6], demonstrating their promise as a source of novel and/or bioactive natural products. Our previous chemical investigation of the bioactive secondary metabolites produced by the endophytic Aspergillus sp. YXf3 associated with Ginkgo biloba led to the isolation of new p-terphenyls and novel types of diterpenoids including pimarane-type diterpenoids (sphaeropsidins A and B, aspergiloids D and E), a cleistanthane-type diterpenoid (aspergiloid C), and norcleistanthane-type diterpenoids (asergiloids A, B, and F–H), many of which were reported from this microorganism for the first time [7-9]. Interestingly, sphaeropsidins A and B were also discovered from both Aspergillus chevalieri and phytopathogenic fungus Sphaeropsis sapinea, displaying anti-gram-positive bacterial, antiviral, antiprotozoal and phytotoxic activity [10-12]. We further focused on the fractionation containing the minor terpenoid constituents with characteristic signals for terminal vinyl group detected by 1H NMR from the liquid fermentation broth of Aspergillus sp. YXf3 and isolated a novel norditerpenoid, namely, aspergiloid I (1) (Figure 1). Herein, we report the production, isolation, structure characterization, and biological activity of 1, a rare spirolactone metabolite with a novel carbon skeleton.
Figure 1: Structure of aspergiloid I (1) and its novel skeleton.
Figure 1: Structure of aspergiloid I (1) and its novel skeleton.
Results and Discussion
A large-scale culture broth of Aspergillus sp. YXf3 was extracted with EtOAc and separated by a combination of column chromatographic methods. A preliminary survey of the fractionation by 1H NMR and LC–MS revealed the presence of the molecule we eventually named aspergiloid I (1), which had a low-resolution mass ([M + Na]+ at m/z 329) spectrum that did not match any previously isolated compounds, prompted us to purify it further. The obtained fermented broth (~45 L) was extracted four times with EtOAc (v/v, 1:1) to afford a brown crude extract (9.1 g). Subsequent fractionation by silica gel column chromatography (CC), octadecylsilyl (ODS) CC, Sephadex LH-20 and semi-preparative reversed-phase HPLC yielded 1 (4.3 mg).
Aspergiloid I (1) was isolated as a colorless lamellar crystal, with the molecular formula C18H26O4 (6 double-bond equivalents) as derived from the ESI high-resolution mass spectrometry ([M + Na]+ at m/z 329.1729, calculated 329.1723) and NMR data (Table 1). The IR spectrum exhibited absorptions at 3649 (hydroxy group) and 1735 cm−1 (carbonyl group). The 1H NMR spectrum (acquired in DMSO-d6) displayed signals of a terminal vinyl group at δH 5.72 (H-14), 4.99 (H-15α) and 4.96 (H-15β), two oxygenated methine protons (δH 3.76, 2.81), ten aliphatic protons [δH 2.56, 2.25, 1.96, 1.63, 1.61, 1.51 (2H), 1.42, 1.20, 1.11], three aliphatic methyl groups (δH 1.03, 1.02, 0.86) and two hydroxy groups (δH 5.21, 5.04). The 13C NMR and DEPT spectra revealed that 1 contained 18 carbons, attributable to three methyl groups (δC 23.4, 24.5, 29.2), five aliphatic methylenes (δC 18.1, 23.7, 28.4, 28.5, 31.8), one olefinic methylene (δC 113.2), two oxygenated methines (δC 76.9, 64.7), one olefinic methine (δC 142.9), and six non-protonated carbon atoms (one of which was identified as lactone group) (δC 35.1, 40.0, 88.6, 126.6, 170.9, 171.6). These data show that 1 has two double bonds and one carbonyl which require three degrees of unsaturation, thus, 1 must also contain three rings.
Table 1: 1H and 13C NMR spectroscopic data for aspergiloid I (1).
| Position | δCa | δHa (mult, J in Hz) | δCb | δHb (mult, J in Hz) |
|---|---|---|---|---|
| 1 | 28.5, CH2 |
1.96, td (13.5, 4.5, Hα);
1.20, br d (14.0, Hβ) |
28.9 |
2.00, td (13.5, 4.0, Hα);
1.43, br d (13.5, Hβ) |
| 2 | 18.1, CH2 |
1.63, m (Hα);
1.51, m (overlap, Hβ) |
18.2 |
1.87, qt (13.5, 4.0, Hα);
1.61, dq (13.5, 4.0, Hβ) |
| 3 | 31.8, CH2 |
1.51, m (overlap, Hα);
1.11, br d (13.5, Hβ) |
32.0 |
1.53, td (13.5, 4.0, Hα);
1.32, br d (13.5, Hβ) |
| 4 | 35.1, C | 35.3 | ||
| 5 | 76.9, CH | 2.81, d (7.0) | 79.0 | 3.09, br s |
| 5-OH | 5.21, d (7.0) | |||
| 6 | 88.6, C | 88.8 | ||
| 7 | 170.9, C | 171.5 | ||
| 8 | 23.7, CH2 |
2.56, ddd (20.0, 13.0, 4.5, Hα);
2.25, dd (20.0, 4.5, Hβ) |
23.7 | 2.78, dtd (20.0, 6.7, 1.5, Hα); 2.34, dtd (20.0, 6.0, 1.5, Hβ) |
| 9 | 28.4, CH2 |
1.61, td (13.0, 6.0, Hα);
1.42, dd (13.0, 6.0, Hβ) |
30.5 | 1.71, tdd (13.5, 7.5, 6.0, Hα); 1.64, dt (13.5, 6.0, Hβ) |
| 10 | 40.0, C | 39.9 | ||
| 11 | 64.7, CH | 3.76, d (6.5) | 67.4 | 4.31, s |
| 11-OH | 5.04, d (6.5) | |||
| 12 | 126.6, C | 126.9 | ||
| 13 | 171.6, C | 172.1 | ||
| 14 | 142.9, CH | 5.72, dd (17.5, 11.0) | 143.7 | 5.80, dd (17.2, 11.0) |
| 15 | 113.2, CH2 |
4.99, dd (17.5, 1.5, Hα);
4.96, dd (11.0, 1.5, Hβ) |
113.1 |
5.06, d (17.2, Hα);
5.05, d (11.0, Hβ) |
| 16 | 23.4, CH3 | 1.02, s | 20.1 | 1.08, s |
| 17 | 24.5, CH3 | 1.03, s | 24.6 | 1.17, s |
| 18 | 29.2, CH3 | 0.86, s | 28.6 | 0.97, s |
aAcquired in DMSO-d6 (125 MHz and 500 MHz). bAcquired in CDCl3 (125 MHz and 500 MHz).
The gross structure of 1 was initially deduced by comprehensive analysis of its 1D and 2D NMR data. The 13C NMR and HSQC spectra of 1 allowed all protons to be assigned to their respective carbons. The 1H,1H three-bond couplings from H-1 to H-3 observed in the COSY experiment established a spin system from C-1 to C-3 (Figure 2). The COSY correlation between H-8 and H-9 revealed C-8 to C-9 connectivity. A terminal vinyl moiety H-14/H2-15 was also confirmed by 1H,1H-COSY correlations. The hydroxy group (δH 5.21) attached to C-5 and the other hydroxy group (δH 5.04) attached to C-11 were identified by the 1H,1H couplings (acquired in DMSO-d6) with H-5 (δH 2.81), and H-11 (δH 3.76), respectively. HMBC correlations from two singlet methyl groups’ protons H3-17 and H3-18 to C-3, C-4, and C-5 indicate that C-17 and C-18 were located on the same quaternary carbon C-4, which was connected by C-3 and C-5. HMBC correlations from the hydroxy proton (δH 5.21) to C-5, and C-6 (acquired in DMSO-d6), and from H-5 to C-1, C-6, and C-7 (acquired in CDCl3) assigned the connectivity of the C-6 to C-1, C-5, and C-7. The other singlet methyl group, C-16, and the terminal vinyl group (C-14–C-15), were also located on the same quaternary carbon, C-10, which was flanked by C-9 and C-11 deduced from the HMBC correlations from H2-9 to C-10, C-11, and C-14, from H-11 to C-14, from H2-15 to C-10, and from the methyl protons (H3-16; δH 1.02) to C-10, C-11, and C-14. The HMBC correlations from the hydroxy proton (δH 5.04) to C-12, from H-11 to C-7, C-12, and C-13 indicated that C-12 was linked to C-7, C-11, and C-13 to form an α,β-unsaturated enone fragment. HMBC correlations from H2-8 to C-7 and C-12 secured the connectivity of the C-8 to C-7. The connectivity of C-6 to the ketone carbon C-13 through an ester linkage, which was also supported by the downfield chemical shift of C-6 (δC 88.6) completed the structure of 6/5/6 tricyclic spirolactone. The relative configuration of aspergiloid I (1) could be determined by NOESY correlations. The NOESY spectrum showed correlations of OH-5 with H3-18 and H-8α, of H-9β with H-8α and H3-16, and of H3-16 with OH-11, indicating that OH-5, H3-16, and OH-11 were on the same plane, while the relative configuration of the chiral center C-6 was further confirmed by a NOESY correlation of H-1α with H-8β. Therefore, the structure of compound 1 was elucidated as shown in Figure 1, representing a new type of carbon skeleton in the norditerpenoid family.
![[1860-5397-10-282-2]](/bjoc/content/figures/1860-5397-10-282-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Selected 1H,1H-COSY and HMBC correlations and key NOEs observed for 1.
Figure 2: Selected 1H,1H-COSY and HMBC correlations and key NOEs observed for 1.
The structure of 1 was further confirmed by a low-temperature (100 K) single-crystal X-ray diffraction experiment, which is shown in Figure 3. As compound 1 has a relatively high percentage of oxygen, it shows enough anomalous dispersion of Cu Kα radiation and allows to determinate the absolute stereochemistry with the Hooft parameter 0.17(15) for 992 Bijvoet pairs by single-crystal X-ray diffraction experiment [13]. Therefore, the absolute configurations of the chiral centers in 1 were established as 5R, 6S, 10R, 11R. This compound is structurally characterized by a new carbon skeleton with an unprecedented 6/5/6 tricyclic ring system bearing an α,β-unsaturated spirolactone moiety in ring B. The skeleton, tentatively named aspergilane, represents a new subclass of norditerpenoids.
![[1860-5397-10-282-3]](/bjoc/content/figures/1860-5397-10-282-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: X-ray single-crystal structure of 1.
Figure 3: X-ray single-crystal structure of 1.
The endophytic fungus Aspergillus sp. YXf3 can produce different types of diterpenoids, including pimarane-type diterpenoids (sphaeropsidin A and B, aspergiloid D and E), cleistanthane type-diterpenoid (aspergiloid C), and norcleistanthane-type diterpenoids (aspergiloid A, B, and F–H) [7-9], and the “aspergilane”-type norditerpenoid aspergiloid I. A plausible biogenetic relation is given in Scheme S1 (Supporting Information File 1) for the formation of these diterpenoids. Here the hypothetical pimarane compound 2, the hemiketal lactone ring-opening product of aspergiloid E, was proposed as the most probable biosynthetic intermediate. As shown in Scheme 1, we suggest the biosynthesis of 1 starts from the classical diterpene precursor geranylgeranyl diphosphate [14], and intermediate 2 undergoes decarboxylation to form 3 through Baeyer–Villiger oxidation to form the 7-membered lactone 4, then hydrolyzation, decarboxylation and lactonization to finally give aspergiloid I (1).
Scheme 1: Proposed biosynthetic pathway of 1.
Scheme 1: Proposed biosynthetic pathway of 1.
Aspergiloid I (1) was evaluated for its cytotoxicity against eleven human cancer cell lines, K562 myeloid leukemia, SH-SY5Y neuroblastoma, SGC-7901 gastric adenocarcinoma, HepG2, SMMC-7721 hepatocellular carcinoma, A549 lung cancer, MCF-7, MDA-MB-231 breast cancer, HCT116, SW480 colon cancer, HT29 colorectal cancer. However, no significant activity was detected (IC50 > 50 μM). It also displayed no anti-oxidant, and acetylcholinesterase (AChE), α-glucosidase, and topoisomerase IIα inhibitory activities at a concentration of 50 μg/mL. Antimicrobial activities against a variety of plant pathogenic bacteria (Xanthomonas oryzae pv. oryzae Swings, Xanthomonas oryzae pv. oryzicola Swings, Acidovorax avenae subsp. Citrulli, Erwinia amylovora, Pseudomonas syringae pv. Lachrymans, Clavibacter michiganense subsp. Sepedonicus, and Pectobacterium carotovorum subsp. carotovorum) and fungi (Rhizoctonia solani Kühn, Rhizotonia cerealis van der Hoeven, Phytophthora capsici Leonian, Fusarium moniliforme Sheld, Alternaria solani Jones et Grout, Sclerotinia sclerotiorum de Bary, Fusarium graminearum Schw., Fusarium coeruleum Sacc., and Botrytis cinerea Pers.) were tested. Aspergiloid I (1) showed no antibacterial or antifungal activity at a concentration of 20 μg/mL. It also had no antifungal activity against Candida albicans (ATCC 10231) or Fusarium oxysporum f. sp. cubense race 4 at concentrations as high as 20 mg/mL.
Conclusion
In summary, guided by 1H NMR detection, we isolated and characterized a novel norditerpenoid, aspergiloid I (1), from the liquid fermentation broth of the endophytic Aspergillus sp. YXf3 associated with Ginkgo biloba. This compound is structurally characterized by a new carbon skeleton with an unprecedented 6/5/6 tricyclic ring system bearing a α,β-unsaturated spirolactone moiety, and represents a new subclass of norditerpenoid, the skeleton of which is named aspergilane. Chemical investigation of Aspergillus sp. YXf3 revealed it is a pluripotent fungus which can produce different types of novel interesting metabolites, including p-terphenyls, pimarane, cleistanthane, norcleistanthane-type diterpenoids [7], and the “aspergilane”-type norditerpenoid 1. It is possible to propose that 1 is biosynthetically derived from hypothetical intermediate pimarane compound 2, the hemiketal lactone ring-opening product of aspergiloid E. In biological tests, 1 showed no cytotoxic, antimicrobial, anti-oxidant, acetylcholinesterase (AChE), α-glucosidase, and topoisomerase IIα inhibitory activities. In order to perform more biological assays for this unusual norditerpenoid, further scale-up isolation is in progress.
Experimental
General experimental procedures
The melting point was measured on a Beijing Taike X-5 stage apparatus and reported without correction. The optical rotation was recorded using a Rudolph Autopol III polarimeter. The UV spectrum was obtained on a Hitachi U-3000 spectrophotometer. The CD spectrum was measured on a JASCO J-810 spectrometer, and the IR spectrum (KBr) was obtained on a Nexus 870 FTIR spectrometer. NMR data were acquired using a Bruker AVANCE III-500 NMR spectrometer at 500 MHz for 1H NMR and 125 MHz for 13C NMR. The chemical shifts were given in δ (ppm) and referenced to the solvent signal (DMSO-d6, δH 2.50, δC 39.5; CDCl3, δH 7.26, δC 77.1) as the internal standard, and coupling constants (J) are reported in Hz. The high resolution mass measurement was conducted on an Agilent 6210 TOF LC–MS spectrometer. Silica gel (200–300 mesh; Qingdao Marine Chemical Factory, Qingdao, China) and Sephadex LH-20 gel (Pharmacia Biotech, Sweden) were used for column chromatography (CC). Semipreparative HPLC was conducted on a Waters ODS (250 × 4.6 mm, 5 μm) on a Hitachi HPLC system consisting of a L-7110 pump (Hitachi) and a L-7400 UV–vis detector (Hitachi). All other chemicals used in this study were of analytical grade.
Fungal material, cultivation, extraction and isolation
The fungal strain Aspergillus sp. YXf3 was isolated by one of the authors (Z.K.G.) from a healthy leaf of Ginkgo biloba collected in the campus of Nanjing University (Nanjing, P. R. China), in October 2008 [7]. The strain was cultured on MEA (consisting of 20 g/L malt extract, 20 g/L sucrose, 1 g/L peptone, 20 g/L agar and deionized water) at 28 °C for 5 days. Agar plugs were used to inoculate in 1000 mL Erlenmeyer flasks, each containing 300 mL of ME liquid media. Fermentation was carried out on a rotary shaker (140 rpm) at 26 °C for 13 days in 1000 mL Erlenmeyer flasks. Mycelia were separated by filtration and the obtained fermented broth (about 45 L) was extracted four times with EtOAc (v/v, 1:1) to afford a brown crude extract (9.1 g), which was then fractionated by silica gel (91 g) CC (8 × 100 cm) eluted with a gradient of CHCl3–MeOH (v/v 100:0, 100:1, 100:2, 100:4, 100:8, 100:16, 0:100, each 1200 mL) to produce seven fractions. Fraction 2 (1.52 g; CHCl3–MeOH, 100:1) was separated on a ODS column (5 × 50 cm) with a gradient of MeOH–H2O (v/v 30:70, 40:60, 50:50, 60:40, 70:30, 80:20, 100:0, each 500 mL) to give seven subfractions. The fourth subfraction (108.6 mg; MeOH–H2O, 60:40) was further isolated by Sephadex LH-20 CC eluting with MeOH and purified by semipreparative reversed-phase HPLC to yield 1 (4.3 mg) (64% MeOH in water; tR = 17.94 min).
Aspergiloid I (1): colorless lamellar crystals; mp 181.0–184.9 °C; [α]28D −19.5 (c 0.2, MeOH); UV (MeOH) λmax (log ε): 220.0 (3.97) nm; CD (MeOH): Δε = 214 (+5.30), 242 (−8.23), 283 (+0.04), 374 (−0.54), 399 (+0.50), 411 (−0.14), 471 (+1.69), 488 (−0.53), 500 (+1.19) nm; IR (KBr) νmax 3649, 1735, 1700, 1652, 1558, 1540, 1508, 1457, 816 cm−1; 1H and 13C NMR data, see Table 1; HRMS–ESI (m/z): [M + Na]+ calcd for C18H26O4Na, 329.1723; found, 329.1729.
X-ray crystallographic analysis of 1
Colorless crystal of 1 was obtained by crystallizing from a solution of 2 mL methanol with two drops of distilled water. The single crystal X-ray diffraction data were collected at 100 K with Cu Kα radiation (λ = 1.54178 Å) on a Bruker APEX DUO CCD diffractometer, equipped with an Oxford Cryostream 700+ cooler. Structures were solved using the program SHELXS-97 [15], and refined anisotropically by full-matrix least-squares on F2 using SHELXL-97. The absolute configurations were determined by computation of the Hooft parameter [13], in all cases yielding a probability of 1.000 that the reported configuration is correct. Crystal data: C18H26O4, M = 306.39, orthorhombic, a = 6.3349(2) Å, b = 11.1090(3) Å, c = 23.3643(6) Å, α = 90.00°, β = 90.00°, γ = 90.00°, V = 1644.25(8) Å3, T = 100(2) K, space group P212121, Z = 4, μ(Cu Kα) = 0.694 mm−1, 7387 reflections measured, 2729 independent reflections (Rint = 0.0603). The final R1 values were 0.0901 (I > 2σ(I)). The final wR(F2) values were 0.2603 (I > 2σ(I)). The final R1 values were 0.1025 (all data). The final wR(F2) values were 0.2816 (all data). The goodness of fit on F2 was 1.126. The Hooft parameter is 0.17(15) for 992 Bijvoet pairs. Crystallographic data for the structure of aspergiloid I (1) have been deposited with the Cambridge Crystallographic Data Centre (deposition no. CCDC 985728). These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/data_request/cif, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB21EZ, UK; fax: +44-1223-336-033; or desposit@ccdc.cam.ac.uk.
Biological assays
Cytotoxic activity against 11 human cancer cell lines, K562 myeloid leukemia, SH-SY5Y beuroblastoma, SGC-7901 gastric adenocarcinoma, HepG2, SMMC-7721 hepatocellular carcinoma, A549 lung cancer, MCF-7, MDA-MB-231 breast cancer, HCT116, SW480 colon cancer, HT29 colorectal cancer, were evaluated with the MTT assay [16,17]. Antimicrobial activities against a variety of plant pathogenic bacteria (Xanthomonas oryzae pv. oryzae Swings, Xanthomonas oryzae pv. oryzicola Swings, Acidovorax avenae subsp. Citrulli, Erwinia amylovora, Pseudomonas syringae pv. Lachrymans, Clavibacter michiganense subsp. Sepedonicus, and Pectobacterium carotovorum subsp. carotovorum) and fungi (Rhizoctonia solani Kühn, Rhizotonia cerealis van der Hoeven, Phytophthora capsici Leonian, Fusarium moniliforme Sheld, Alternaria solani Jones et Grout, Sclerotinia sclerotiorum de Bary, Fusarium graminearum Schw., Fusarium coeruleum Sacc., Botrytis cinerea Pers., and Fusarium oxysporum f. sp. cubense race 4), and Candida albicans (ATCC 10231), and the antioxidant, acetylcholinesterase (AChE) , α-glucosidase, and topoisomerase IIα inhibitory activities were performed in accordance with the primary literature [18-20].
Supporting Information
| Supporting Information File 1: 1D, 2D NMR spectra, HRMS–ESI, and the X-ray crystallographic structure of 1. | ||
| Format: PDF | Size: 3.2 MB | Download |
Acknowledgements
We thank Dr. Jeffrey Rudolf (The Scripps Resarch Institute, Jupiter, FL) for editing this manuscript. This work was cofinanced by National Natural Science Foundation of China (81172948, 81121062, 21132004, and 41406083), 863 Project (2013AA092903), Project for New Century Excellent Talents in University (NCET-10-0477), Hainan Natural Science Foundation (313079 and 314176), and National Nonprofit Institute Research Grant of CATAS-ITBB (ITBB140405).
References
-
Tan, R. X.; Zou, W. X. Nat. Prod. Rep. 2001, 18, 448–459. doi:10.1039/b100918o
Return to citation in text: [1] -
Zhang, H. W.; Song, Y. C.; Tan, R. X. Nat. Prod. Rep. 2006, 23, 753–771. doi:10.1039/b609472b
Return to citation in text: [1] -
Wang, X.-N.; Bashyal, B. P.; Wijeratne, E. M. K.; U’Ren, J. M.; Liu, M. X.; Gunatilaka, M. K.; Arnold, A. E.; Gunatilaka, A. A. L. J. Nat. Prod. 2011, 74, 2052–2061. doi:10.1021/np2000864
Return to citation in text: [1] -
Sun, H.-F.; Li, X.-M.; Meng, L.; Cui, C.-M.; Gao, S.-S.; Li, C.-S.; Huang, C.-G.; Wang, B.-G. J. Nat. Prod. 2012, 75, 148–152. doi:10.1021/np2006742
Return to citation in text: [1] -
Ding, L.; Dahse, H.-M.; Hertweck, C. J. Nat. Prod. 2012, 75, 617–621. doi:10.1021/np2008544
Return to citation in text: [1] -
Amand, S.; Langenfeld, A.; Blond, A.; Dupont, J.; Nay, B.; Prado, S. J. Nat. Prod. 2012, 75, 798–801. doi:10.1021/np2009913
Return to citation in text: [1] -
Guo, Z. K.; Yan, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. J. Nat. Prod. 2012, 75, 15–21. doi:10.1021/np200321s
Return to citation in text: [1] [2] [3] [4] -
Yan, T.; Guo, Z. K.; Jiang, R.; Wei, W.; Wang, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. Planta Med. 2013, 79, 348–352. doi:10.1055/s-0032-1328260
Return to citation in text: [1] [2] -
Guo, Z. K.; Liu, S. B.; Ma, S. Nat. Prod. Res. Dev. 2013, 25, 778–781.
Return to citation in text: [1] [2] -
Ellestad, G. A.; Kunstmann, M. P.; Mirando, P.; Morton, G. O. J. Am. Chem. Soc. 1972, 94, 6206–6208. doi:10.1021/ja00772a054
Return to citation in text: [1] -
Evidente, A.; Sparapano, L.; Fierro, O.; Bruno, G.; Giordano, F.; Motta, A. Phytochemistry 1997, 45, 705–713. doi:10.1016/S0031-9422(97)00006-X
Return to citation in text: [1] -
Evidente, A.; Sparapano, L.; Bruno, G.; Motta, A. Phytochemistry 2002, 59, 817–823. doi:10.1016/S0031-9422(02)00015-8
Return to citation in text: [1] -
Hooft, R. W. W.; Straver, L. H.; Spek, A. L. J. Appl. Crystallogr. 2008, 41, 96–103. doi:10.1107/S0021889807059870
Return to citation in text: [1] [2] -
Dewick, P. M. Nat. Prod. Rep. 2002, 19, 181–222. doi:10.1039/B002685I
Return to citation in text: [1] -
Sheldrick, G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, 64, 112–122. doi:10.1107/S0108767307043930
Return to citation in text: [1] -
Mosmann, T. J. Immunol. Methods 1983, 65, 55–63.
Return to citation in text: [1] -
Guo, Z. K.; Wang, T.; Guo, Y.; Song, Y. C.; Tan, R. X.; Ge, H. M. Planta Med. 2011, 77, 2057–2060. doi:10.1055/s-0031-1280097
Return to citation in text: [1] -
Yang, D. L.; Mei, W. L.; Wang, H.; Dai, H. F. Z. Naturforsch., B 2010, 65, 757–761.
Return to citation in text: [1] -
Zhang, Z.; Liao, L.; Moore, J.; Wu, T.; Wang, Z. Food Chem. 2009, 113, 160–165. doi:10.1016/j.foodchem.2008.07.061
Return to citation in text: [1] -
Ellman, G. L.; Courtney, K. D.; Andres, V., Jr.; Featherstone, R. M. Biochem. Pharmacol. 1961, 7, 88–95. doi:10.1016/0006-2952(61)90145-9
Return to citation in text: [1]
| 1. | Tan, R. X.; Zou, W. X. Nat. Prod. Rep. 2001, 18, 448–459. doi:10.1039/b100918o |
| 2. | Zhang, H. W.; Song, Y. C.; Tan, R. X. Nat. Prod. Rep. 2006, 23, 753–771. doi:10.1039/b609472b |
| 13. | Hooft, R. W. W.; Straver, L. H.; Spek, A. L. J. Appl. Crystallogr. 2008, 41, 96–103. doi:10.1107/S0021889807059870 |
| 10. | Ellestad, G. A.; Kunstmann, M. P.; Mirando, P.; Morton, G. O. J. Am. Chem. Soc. 1972, 94, 6206–6208. doi:10.1021/ja00772a054 |
| 11. | Evidente, A.; Sparapano, L.; Fierro, O.; Bruno, G.; Giordano, F.; Motta, A. Phytochemistry 1997, 45, 705–713. doi:10.1016/S0031-9422(97)00006-X |
| 12. | Evidente, A.; Sparapano, L.; Bruno, G.; Motta, A. Phytochemistry 2002, 59, 817–823. doi:10.1016/S0031-9422(02)00015-8 |
| 7. | Guo, Z. K.; Yan, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. J. Nat. Prod. 2012, 75, 15–21. doi:10.1021/np200321s |
| 8. | Yan, T.; Guo, Z. K.; Jiang, R.; Wei, W.; Wang, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. Planta Med. 2013, 79, 348–352. doi:10.1055/s-0032-1328260 |
| 9. | Guo, Z. K.; Liu, S. B.; Ma, S. Nat. Prod. Res. Dev. 2013, 25, 778–781. |
| 18. | Yang, D. L.; Mei, W. L.; Wang, H.; Dai, H. F. Z. Naturforsch., B 2010, 65, 757–761. |
| 19. | Zhang, Z.; Liao, L.; Moore, J.; Wu, T.; Wang, Z. Food Chem. 2009, 113, 160–165. doi:10.1016/j.foodchem.2008.07.061 |
| 20. | Ellman, G. L.; Courtney, K. D.; Andres, V., Jr.; Featherstone, R. M. Biochem. Pharmacol. 1961, 7, 88–95. doi:10.1016/0006-2952(61)90145-9 |
| 3. | Wang, X.-N.; Bashyal, B. P.; Wijeratne, E. M. K.; U’Ren, J. M.; Liu, M. X.; Gunatilaka, M. K.; Arnold, A. E.; Gunatilaka, A. A. L. J. Nat. Prod. 2011, 74, 2052–2061. doi:10.1021/np2000864 |
| 4. | Sun, H.-F.; Li, X.-M.; Meng, L.; Cui, C.-M.; Gao, S.-S.; Li, C.-S.; Huang, C.-G.; Wang, B.-G. J. Nat. Prod. 2012, 75, 148–152. doi:10.1021/np2006742 |
| 5. | Ding, L.; Dahse, H.-M.; Hertweck, C. J. Nat. Prod. 2012, 75, 617–621. doi:10.1021/np2008544 |
| 6. | Amand, S.; Langenfeld, A.; Blond, A.; Dupont, J.; Nay, B.; Prado, S. J. Nat. Prod. 2012, 75, 798–801. doi:10.1021/np2009913 |
| 7. | Guo, Z. K.; Yan, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. J. Nat. Prod. 2012, 75, 15–21. doi:10.1021/np200321s |
| 13. | Hooft, R. W. W.; Straver, L. H.; Spek, A. L. J. Appl. Crystallogr. 2008, 41, 96–103. doi:10.1107/S0021889807059870 |
| 7. | Guo, Z. K.; Yan, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. J. Nat. Prod. 2012, 75, 15–21. doi:10.1021/np200321s |
| 16. | Mosmann, T. J. Immunol. Methods 1983, 65, 55–63. |
| 17. | Guo, Z. K.; Wang, T.; Guo, Y.; Song, Y. C.; Tan, R. X.; Ge, H. M. Planta Med. 2011, 77, 2057–2060. doi:10.1055/s-0031-1280097 |
| 7. | Guo, Z. K.; Yan, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. J. Nat. Prod. 2012, 75, 15–21. doi:10.1021/np200321s |
| 8. | Yan, T.; Guo, Z. K.; Jiang, R.; Wei, W.; Wang, T.; Guo, Y.; Song, Y. C.; Jiao, R. H.; Tan, R. X.; Ge, H. M. Planta Med. 2013, 79, 348–352. doi:10.1055/s-0032-1328260 |
| 9. | Guo, Z. K.; Liu, S. B.; Ma, S. Nat. Prod. Res. Dev. 2013, 25, 778–781. |
| 15. | Sheldrick, G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, 64, 112–122. doi:10.1107/S0108767307043930 |
© 2014 Guo et al; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (http://www.beilstein-journals.org/bjoc)