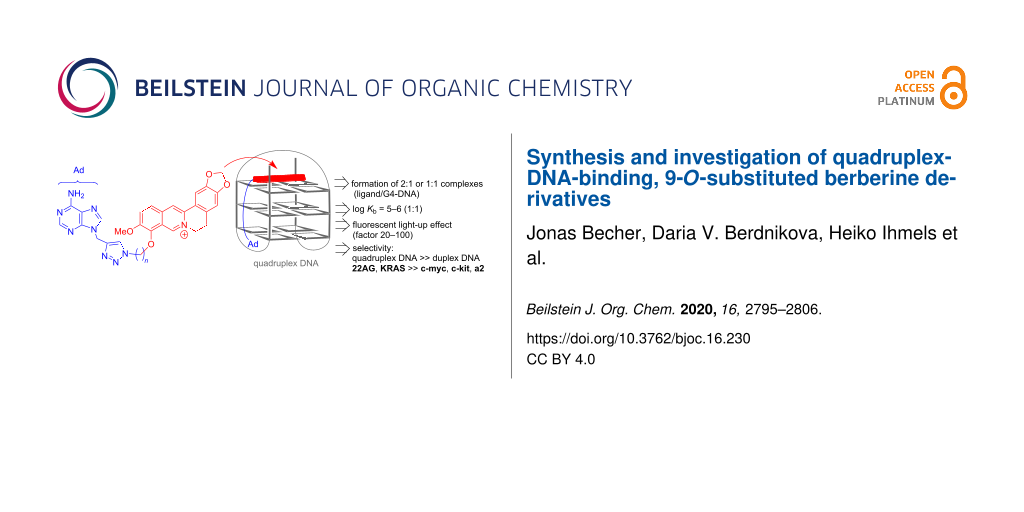
Abstract
A small series of five novel berberine derivatives was synthesized by the Cu-catalyzed click reaction of 9-propargyladenine with 9-O-(azidoalkyl)berberine derivatives. The association of the resulting berberine–adenine conjugates with representative quadruplex-forming oligonucleotides 22AG dA(G3TTA)3G3 and a2 d(ACAG4TGTG4)2 was examined with photometric and fluorimetric titrations, thermal DNA denaturation analysis, and CD spectroscopy. The results from the spectrometric titrations indicated the formation of 2:1 or 1:1 complexes (ligand:G4-DNA) with log Kb values of 10–11 (2:1) and 5–6 (1:1), which are typical for berberine derivatives. Notably, a clear relationship between the binding affinity of the ligands with the length of the alkyl linker chain, n, was not observed. However, depending on the structure, the ligands exhibited different effects when bound to the G4-DNA, such as fluorescent light-up effects and formation of ICD bands, which are mostly pronounced with a linker length of n = 4 (with a2) and n = 5 (with 22AG), thus indicating that each ligand–G4-DNA complex has a specific structure with respect to relative alignment and conformational flexibility of the ligand in the binding site. It was shown exemplarily with one representative ligand from the series that such berberine–adenine conjugates exhibit a selective binding, specifically a selectivity to quadruplex DNA in competition with duplex DNA, and a preferential thermal stabilization of the G4-DNA forms 22AG and KRAS. Notably, the experimental data do not provide evidence for a significant effect of the adenine unit on the binding affinity of the ligands, for example, by additional association with the loops, presumably because the adenine residue is sterically shielded by the neighboring triazole unit.
Graphical Abstract
Introduction
In nucleic acids chemistry, quadruplex DNA (G4-DNA) has been established as an attractive target [1-3]. This noncanonical DNA form is assembled through stacking of at least two guanine quartets and has been observed with highly diverse variation of structures in guanine-rich DNA sequences [4-6], for example, in the promoter regions of oncogenes or in single-stranded overhang of telomeric DNA [7-9]. Most notably, it has been shown that quadruplex formation is directly involved in biologically relevant processes [10], for example, in the suppression of gene expression [11,12] or the induction of the cellular response to DNA damage [13,14]. Because of the increasing evidence of an essential biological function of G4-DNA, this DNA form is considered as an attractive target in drug development [1,2,15,16]. For that purpose, G4-DNA-targeting ligands are searched for that bind selectively and sufficiently strong to quadruplex DNA and thereby influence the biological function of G-rich DNA sequences [17-22]. Among the numerous classes of compounds, mostly related to traditional DNA binders, that have been successfully developed as G4-DNA ligands [17], the natural product berberine (1a) has attracted special attention. Berberine (1a) is an isoquinoline alkaloid with an exceptionally wide range of biological activities [23,24]. It has been shown that berberine (1a) and its derivatives act, for example, as anti-inflammatory [25], antibacterial [26,27], and anticancer reagents [28,29]. The latter property is mainly based on the binding interaction of berberine with nucleic acids and the resulting inhibition of topoisomerase and telomerase [2,30]. Most notably, berberine (1a) induces a strong growth inhibition in several human cancer cells, but has only a relatively low cytotoxicity in healthy cells [31,32]. Berberine is also known as a G4-DNA ligand [33]. Especially berberine derivatives that carry additional substituents with varying alkyl chain lengths in the 9- and 13-position show enhanced binding properties and high selectivity towards telomeric G-quadruplex DNA [34-37]. Representative examples of this class of compounds are the 9-O-aminoalkyl-substituted and 9-O-pyridinium-N-alkyl-substituted derivatives 1bn and 1cn or the 13-phenylalkyl-substituted substrates 1dn or 1en (Scheme 1) [38-42]. In the latter cases, the binding properties depend on the length of the alkyl chain. For example, the aminohexyl-substituted derivative 1b6 and the phenylpropyl-substituted compound 1d3 have the highest affinity to G-quadruplex DNA, whereas the derivatives with other alkyl chain lengths have a lower affinity [38]. Along the same line, the influence of the length and substituents of the side chains at the G4-DNA ligands have been assessed for quinolinium [43], indoloquinoline [44,45], phenanthroline [46], phenothiazine [47], and thiazole orange [48] derivatives. In these studies, the delicate balance between the hydrophobic effects of the alkyl chain and the thermodynamically favorable interactions on the association of ammonium or pyridinium groups in the grooves and loops was assessed. In another approach with a cyanine-based ligand, the alkyl substituents with a suitable length were terminated with an N-benzylamide functionality to establish the attractive hydrogen bonding and π stacking with the thymidine residues in the loops in G4-DNA, so that this ligand binds with very high selectivity to the particular quadruplex-forming oligonucleotide J19 [49].
Scheme 1: The structures and numbering of berberine (1a) and the alkyl-substituted derivatives 1an–en and the binding equilibrium with quadruplex DNA (G4-DNA).
Scheme 1: The structures and numbering of berberine (1a) and the alkyl-substituted derivatives 1an–en and the...
Overall, the above-mentioned observations indicate that berberine is among the more promising lead structures for the development of G4-DNA ligands. Moreover, the systematic variation of functional side-chains appears to be a suitable approach to determine the factors that influence the selectivity and affinity of a given ligand system. With this background, we proposed that the functionalization of the berberine scaffold with adenine-appended alkyl substituents may provide a useful platform to further explore this important aspect. Specifically, we wished to examine whether adenine–berberine conjugates with a varying linker length may allow to deduce a relationship between the chain length and the binding properties. The adenine unit was supposed to establish binding interactions with the loop region of the quadruplex, namely through Watson–Crick base pairing with the complementary thymidine residues. Herein, we describe the synthesis and characterization of the novel berberine–adenine conjugates 4a–e along with the preliminary investigations of the interactions with selected G4-DNA forms, mainly 22AG as the representative telomeric DNA sequence that may be considered a well-established reference, and a2, i.e., a quadruplex-forming repeat unit from the “insulin-linked polymorphic region” (ILPR) [50], that was also shown to bind quadruplex ligands [51].
Results
Synthesis
As the Cu-catalyzed click reaction between azides and alkynes is a well-established method for the variable functionalization of G4-DNA ligands [52], the berberine–adenine conjugates 4a–e were synthesized by the reaction of 9-propargyladenine (2) [53] with the 9-azidoalkylberberine derivatives 3a–e [54] (Scheme 2). Although the compounds 4a–e formed as the major products in this reaction (>>50%), they were only obtained as isolated products in low to moderate yields (16–38%), mainly because of severe difficulties to completely remove the copper ions that apparently bind tightly to the compounds. The new compounds 4a–e were identified and fully characterized with NMR spectroscopy (1H, 13C, COSY, HSQC, HMBC), mass spectrometry, and elemental analysis.
Scheme 2: Synthesis of the berberine–adenine conjugates 4a–e.
Scheme 2: Synthesis of the berberine–adenine conjugates 4a–e.
DNA-binding properties
Spectrometric titrations
The interactions of the conjugates 4a–e with the quadruplex-forming oligonucleotides 22AG dA(G3TTA)3G3 and a2 d(ACAG4TGTG4)2 were analyzed by photometric and fluorimetric titrations (Figure 1). In all cases, the initial absorption maxima of the ligands 3a–e decreased upon the addition of 22AG or a2 and new, slightly red-shifted absorption bands developed (Figure 1A; cf. Supporting Information File 1, Figure S1). During most of the titrations, the formation of an isosbestic point was observed. However, in several cases it clearly faded away at the end of the titration.
![[1860-5397-16-230-1]](/bjoc/content/figures/1860-5397-16-230-1.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 1: Representative spectrophotometric (A) and spectrofluorimetric (B) titration of compound 4c with 22AG (1) and a2 (2) (c3c = 20 μM; cDNA = 200 μM) in K+-phosphate buffer (pH 7.0) with 10% v/v DMSO). The arrows indicate the development of the absorption or emission bands with increasing DNA concentration. Insets: Plots of Abs./Abs.0 and I/I0, respectively, versus cDNA/c3c. Inset B1: Picture of the emission color and intensity of compound 4c in the absence (left) and the presence (right) of 22AG.
Figure 1: Representative spectrophotometric (A) and spectrofluorimetric (B) titration of compound 4c with 22AG...
The compounds 4a–e have a very low intrinsic emission intensity that increased significantly upon the addition of the G4-DNA 22AG and a2 (Table 1, Figure 1B; cf. Supporting Information File 1, Figure S2). Thus, the characteristic emission band of berberine at 520 nm developed and the relative intensity, I/Io, increased by factors ranging between 21 and 20 for 4b and 71 and 107 for 4c. This light-up effect can be easily followed by the naked eye (Figure 1B1, inset).
Table 1: The binding constants, Kb, fluorescence light-up factors, I/I0, and shifts of the melting temperature, ΔTm, of compounds 4a–e with G4-DNA.
| log Kba (ligand/DNA) | I/I0b | log Kba (ligand/DNA) | I/I0b | ΔTm [°C]c | ΔTm [°C]c | ||||
| 22AG | a2 | F21T | Fa2T | ||||||
| 4a | 5.89 ± 0.07 (1:1) | 39 | 5.27 ± 0.08 (1:1), 10.3 ± 0.1 (2:1) | 58 | 3.4 | −0.5 | |||
| 4b | 5.64 ± 0.15 (1:1), 10.8 ± 0.2 (2:1) | 21 | 5.39 ± 0.14 (1:1), 10.6 ± 0.2 (2:1) | 20 | 6.2 | 0.9 | |||
| 4c | 5.23 ± 0.09 (1:1), 10.7 ± 0.1 (2:1) | 107 | 10.6 ± 0.3 (2:1) | 71 | 6.4 | −0.1 | |||
| 4d | 5.09 ± 0.13 (1:1), 10.8 ± 0.1 (2:1) | 94 | 11.1 ± 0.2 (2:1) | 52 | 9.9 | 0.0 | |||
| 4e | 10.7 ± 0.1 (2:1) | 29 | 11.1 ± 0.1 (2:1) | 35 | 12.9 | 1.1 | |||
aDetermined from the analysis of the photometric titration data with Specfit/32TM with the adequate fits for complexes with ligand:DNA ratio 1:1 and 2:1. K in M−1 for 1:1 complexes and M−2 for 1:2 complexes. bDetermined from the fluorimetric titrations. cDetermined from the fluorimetric FRET experiment of F21T or Fa2T (cDNA = 0.2 µM) at LDR 5.0 in Na-cacodylate buffer [c(K+) = 10 mM, pH 7.2]; LDR = 5; estimated error ± 0.5 °C.
The data from the photometric titrations were used to construct the corresponding binding isotherms and to determine the binding constants, Kb, of 4a–e with G4-DNA 22AG and a2 (cf. Supporting Information File 1). As a general trend, the experimental data could be adequately fitted to a binding stoichiometry ligand/G4-DNA of 2:1 or 1:1. Except for compound 4a, all ligands formed 2:1 complexes with 22AG and a2. The complexes of ligands 4b–e with 22AG have essentially the same log Kb values at 10.7–10.8 (Kb in M−2), whereas the log Kb values of ligands 4a–e with a2 increase slightly in the 2:1 complexes from 10.3 to 11.1 with increasing chain length n (Table 1). At the same time, 1:1 complexes were found for ligands 4a–d and 22AG as well as for ligands 4a,b and a2 with log Kb values between 5.1 (4d and 22AG) and 5.9 (4a and 22AG).
Thermal DNA denaturation analysis
In addition, the thermal stabilization of the G4-DNA 22AG and a2 upon the binding of the ligands 4a–e was investigated by thermal DNA denaturation experiments. For that purpose, the DNA melting temperature Tm of the dye-labeled oligonucleotides F21T and Fa2T (for sequence see caption of Figure 2) was monitored by fluorescence spectroscopy, as the thermally induced unfolding of the quadruplex disrupts the Förster resonance energy transfer (FRET) between the two dyes. With this assay, the thermodynamic stabilization or destabilization of the quadruplex structure upon the complexation of the ligand is indicated by the shift of the melting temperature ΔTm. The analysis revealed an increasing stabilization of the quadruplex F21T toward dissociation with rising concentration of the ligand and with increasing chain length n of the ligands 4a–e, as indicated by the shifts of the melting temperature of up to ΔTm = 12.9 °C (Table 1). In contrast, the oligonucleotide Fa2T is only stabilized to a negligible extent upon the association of the ligands 4a–e (Table 1; cf. Supporting Information File 1, Figure S4). In addition, it was examined exemplarily with the derivative 4e whether the ligand also stabilizes other G4-DNA forms with different topologies. For that purpose, the representative quadruplex-forming oligonucleotides FmycT, FkitT, and FkrasT were also submitted to the thermal DNA denaturation experiments in the presence of 4e (Figure 2; cf. Supporting Information File 1, Figure S5). In all cases, the quadruplex structure is stabilized by the ligand, but it was also observed that the degree of the G4-DNA stabilization upon the binding of the ligand 4e is not the same for the different oligonucleotides as shown by the significantly different ΔTm values. Specifically, the stabilization decreased in the order F21T (ΔTm = 12.9 °C) > FkrasT (ΔTm = 10.1 °C) > FmycT (ΔTm = 5.0 °C) > FkitT (ΔTm = 2.7 °C) > Fa2T (ΔTm = 1.1 °C; at LDR = 5). Furthermore, the selectivity of the ligand 4e toward quadruplex DNA in competition with duplex DNA was investigated by DNA denaturation experiments in the presence of an excess of the duplex-DNA forming oligonucleotide ds26. Under these conditions, the ligand 4e shows essentially the same stabilization as in the absence of ds26 as clearly indicated by only a small decrease of the melting temperature of ΔΔTm = 1.8 °C (Figure 2; cf. Supporting Information File 1, Figure S4).
![[1860-5397-16-230-2]](/bjoc/content/figures/1860-5397-16-230-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Melting temperatures, ΔTm, of G4-DNA (cDNA = 0.2 µM) F21T (black), F21T plus ds26 (15 equiv, red), FkrasT (blue), FmycT (green), FkitT (light blue), and Fa2T (magenta) in the presence of 4e at different LDR = 0.00, 1.25, 2.50, 5.00 in Na-cacodylate buffer (cK+ = 10 mM, pH 7.2); estimated error ±0.5 °C. F21T: fluo-d[(G3TTA)3G3]-tamra; Fa2T: fluo-d[(ACAG4TGTG4)2-tamra; FmycT: fluo-d(TGAG3TG3TAG3TG3TA2)-tamra; FkrasT: fluo-d(AG3CG2TGTG3A2GAG2A)-tamra]; FkitT: fluo-d(AG3AG3CGCTG3AG2AG3)-tamra], fluo = fluorescein, tamra = tetramethylrhodamine; ds26: d(CA2TCG2ATCGA2T2CGATC2GAT2G).
Figure 2: Melting temperatures, ΔTm, of G4-DNA (cDNA = 0.2 µM) F21T (black), F21T plus ds26 (15 equiv, red), ...
CD spectroscopy
The interactions of the ligands 4a–e with G4-DNA 22AG and a2 were also examined with circular dichroism (CD) spectroscopy. Upon the addition of the ligands to 22AG the positive CD band of the DNA at 295 nm remained essentially unchanged, whilst the blue-shifted shoulder to this band disappeared and a negative signal at 260 nm formed, whose intensity depended on the chain length between the berberine and the adenine unit and was the strongest with a chain length of n = 5 (Figure 3). In the case of a2, the positive band of this G4-DNA at 265 nm showed only small fluctuations upon the interaction with the ligands, whereas the intensity of the broad red-shifted shoulder at 295 nm slightly increased at higher LDR. In addition, during all titrations a weak induced CD (ICD) signal was formed in the absorption region of the ligands, which was most pronounced for the ligands with linker lengths of n = 5 on the association with 22AG and of n = 4 on the binding to a2 (Figure 3).
![[1860-5397-16-230-3]](/bjoc/content/figures/1860-5397-16-230-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: CD spectra of 22AG (A) and a2 (B) in the presence of the ligands 4c (in K+-phosphate buffer (pH 7.0 with 10% v/v DMSO) at LDR 0.00 (black), 0.05 (red), 0.20 (blue), 0.50 (magenta), and 1.00 (green); cDNA = 20 µM; T = 20 °C. The arrows indicate the development of the CD bands with increasing LDR. Insets: Plots of intensity of the ICD signal of the ligand–DNA mixtures (LDR 1.0; 361 nm) for 22AG (A) and a2 (B) versus the alkyl chain lengths n of 4a–e.
Figure 3: CD spectra of 22AG (A) and a2 (B) in the presence of the ligands 4c (in K+-phosphate buffer (pH 7.0...
Discussion
The results from the spectrometric titrations with 22AG and a2 clearly revealed the association of the derivatives 4a–e with G4-DNA (Figure 1). Specifically, the red shift and hypochromism of the absorption band along with the pronounced increase of the emission intensity are typical for quadruplex-bound ligands [17-22]. Moreover, the resulting binding constants are in the same range as the ones reported for the resembling derivatives [33-42]. Notably, the analysis of the binding isotherms revealed the formation of 2:1 or 1:1 complexes (ligand/G4-DNA) (Table 1), both of which are also well-established for berberines [33-42]. Based on these general similarities between the derivatives 4a–e and the established quadruplex-binding berberine derivatives it is deduced that the berberine unit in 4a–e binds like the latter ones to the G4-DNA by terminal π-stacking, either with one or two ligands per quadruplex unit, essentially depending on the ligand/DNA ratio in solution.
Although the binding constants of the ligands with 4a–e with 22AG and a2 deviate marginally within the series, they all lie essentially in the same order of magnitude. Thus, clear relationships between the length of the alkyl chain n and the binding constant Kb cannot be deduced from these data, as has been done with other alkyl-substituted berberine derivatives [38-42]. In the latter cases, however, the alkyl chains were substituted with positively charged functionalities that contributed significantly to the binding affinity depending on their spacing from the π-stacking unit. In the case of 4a–e, however, the position of the triazole and adenine unit relative to the berberine does not appear to be highly relevant for the overall binding affinity. It may be concluded that the additional hydrophobic effect is the main contribution of the different substituents of 4a–e to the overall binding affinity.
It is well known that the emission of the parent berberine increases strongly upon the accommodation in sterically constrained binding sites in, e.g., nucleic acids, cucurbiturils, cyclodextrins or micelles [55-58]. Presumably the radiationless deactivation of the excited state by conformational changes, that leads to the low emission intensity in aqueous solution, is suppressed in the sterically restricted binding site. Therefore, it can be deduced that the increased emission of the ligands 4a–e on the addition of G4-DNA is the result of a sufficiently tight complexation. As this fluorescence light-up effect depends significantly on the length of the linker chain n, it is also concluded that the ligands with the strongest effect, i.e., 4c and 4d (n = 4 and 5), have a more restricted molecular flexibility in the binding pocket than the ones with shorter or longer side chains. It should be noted, however, that this binding mode does not lead to a significantly stronger binding affinity as the binding constants of 4c with 22AG or a2 are only slightly different and even smaller than the ones of the other ligands (Table 1).
Obviously, the shifts of the melting temperature ΔTm of G4-DNA in the presence of the ligands do not correlate well with the binding constants. Specifically, none of the ligands stabilizes the quadruplex a2 towards unfolding, as is clearly shown by the negligible shifts of the melting temperatures, whereas the binding constants are in the dimension of 105 M−1. In this context, it has to be emphasized that the binding constants Kb are not directly related to the ligand-induced shifts of the DNA melting temperature Tm, because the latter also depends on other parameters such as binding-site size, cooperativity between ligands, ionic strength, the enthalpy of the DNA denaturation, and on the binding constant and enthalpy of the ligand binding at the melting temperature. However, the binding constant is determined at temperatures below Tm and the enthalpy of the ligand binding (ΔHb) is hardly accessible. Hence, we explain the very low ΔTm values for G4-DNA a2 in the presence of the ligands 4a–e with a very low affinity of these ligands at the melting temperature, which may be caused by the delicate, temperature-dependent equilibrium of the different quadruplex forms of this particular DNA [50,51]. In the case of the G4-DNA 22AG, the stabilization by the ligands 4a–e is more consistent with the binding constants, as both sets of data indicate a moderately high binding constant and a good stabilization towards thermally induced unfolding (Table 1). Nevertheless, the binding constants do not deviate significantly in the series of ligands whereas the thermal stabilization is significantly more pronounced with the ligands 4c and 4d, which may indicate that these ligands have a slightly larger binding affinity to the DNA at the melting temperature than the other ones.
Most notably, the representative DNA denaturation analysis with ligand 4e and different quadruplex forms clearly reveals a significant selectivity. Hence, the hybrid antiparallel G4-DNA 22AG as well as the parallel quadruplex-forming KRAS sequence are stabilized to a significantly more extent than the parallel c-kit, c-myc or the mixed parallel/antiparallel a2 sequence. Therefore, it is concluded that the selectivity of the ligands does not depend on the direction of the strands, i.e., parallel versus antiparallel, but on the loop structure of the respective quadruplex form. In particular, the G4-DNA 22AG and KRAS apparently provide a suitable combination of accessibility of the terminal quartet for π-stacking with a loop structure that enables a favorable accommodation of the side chains. Although it may be assumed that additional interactions of the adenine residue with the loops assist the binding to the loops, there is no clear experimental evidence for this binding mode. This observation is in contrast to the report about an arylalkyl-substituted cyanine dye, that has been shown to bind with a high selectivity to particular G4-DNA forms because of additional attractive interactions with the loops [49]. In the latter case, however, the quadruplex-binding cyanine unit has been proposed to bind in the groove. In this binding mode, the alkyl-appended aryl functionalities may reach the loops and establish additional binding interactions more efficiently than the substituents of terminally stacked quadruplex ligand such as 4a–e.
Along with the selective stabilization of particular quadruplex forms, the DNA denaturation analysis with ligand 4e also showed a high selectivity for the quadruplex stabilization relative to duplex DNA as is clearly shown by only a small decrease of quadruplex melting in the presence of 4e and an excess of the potentially competitive duplex DNA ds26. Although this experiment was only performed exemplarily with the ligand 4e it may be carefully deduced that this class of compounds has a significantly higher affinity to quadruplex DNA.
Additional information about the complex formation between the ligands and the quadruplex DNA forms 22AG and a2 was provided by CD spectroscopy. The changes of the CD spectrum of 22AG upon the addition of derivatives 4a–e clearly indicate a shift of the equilibrium between the different quadruplex forms of 22AG that are formed in the K+-containing buffer solution [59]. In particular, the decrease of the positive shoulder around 270 nm shows the disappearance of the (3 + 1) conformer, to which this band is assigned [59], in favor of the basket-type antiparallel quadruplex structure, which is identified by the characteristic CD pattern with a strong positive band at 295 nm and a weak negative one at 260 nm [60]. These observations show that all ligands stabilize preferentially the basket-type quadruplex structure of 22AG. In the case of the G4-DNA a2 the addition of the ligands shifts the equilibrium between the parallel and antiparallel quadruplex form only slightly in favor of the parallel structure, as is shown by a small increase of the positive CD signal at 295 nm, that is assigned to the parallel quadruplex [61,62], along with a decrease of the CD signal of the antiparallel form at 265 nm (Figure 3).
Notably, weak, but significant ICD bands were observed in the absorption range of the ligands. Such ICD signals of DNA binders result from the dipole–dipole coupling of the ligands with the DNA bases and are typically observed for duplex DNA ligands [63-65]. In the case of the G4-DNA ligands, however, only very weak or even no ICD signals are often observed for the bound molecules, specifically for ligands that bind by terminal π-stacking. To the best of our knowledge, this phenomenon has not been discussed extensively in the literature, so far. As a result, clear relationships between the sign and pattern of the ICD signal of a quadruplex ligand in orientation relative to the binding site, as is well established for duplex binders [63], is not available for quadruplex ligands, yet. With most quadruplex-bound berberine and berberine derivatives, including compounds 1a–d, ICD bands are not formed [34,36,40,66] or at least not explicitly mentioned; however, negative ICD bands [38,67] and exciton-type ICD signals [68] have been reported for some quadruplex-bound berberine derivatives. Unfortunately, in none of the latter cases the ICD signals were directly related to a particular binding mode. However, it was shown by X-ray diffraction analysis of the G4-DNA-bound ligand 1e3 that the berberine unit binds to quadruplex DNA by π-stacking with the 3’-end quartet in a similar mode as the parent berberine; however, in the case of 1e3 the aryl-substituents of the side chain are involved in the additional π-stacking with the G quartet (Figure 4) [38]. At the same time, the ligand 1e3 results a negative ICD when bound to quadruplex. Thus, considering that the derivatives 4a–e have the same berberine fragment as binding unit and assuming that the phase of the ICD signal correlates directly with the relative alignment of the transition dipole moments of the ligand and the DNA bases [64], we carefully conclude that the positive ICD signal of the derivatives 4a–e results from a binding mode in which the berberine unit is in a position essentially perpendicular to the one observed with 1e3 (Figure 4). In this structure the adenylalkyl substituent may be accommodated in the grooves or loops. The latter assumption is somehow supported by the observation that the intensity of the ICD signals varies depending on the chain lengths, indicating that the different fit of the side chain to the groove or loops has a direct influence on the strength and mode of the terminal π-stacking of the berberine unit. The tighter binding with better fitting of the side chain to the binding site is further supported by the observation that both ICD and the fluorescence light-up effect are the strongest with the chain length of 4 and 5 (Table 1).
Figure 4: The simplified structure of the complex between 1e3 and quadruplex DNA (left; [38]) and the proposed orientation of the ligands 4a–e with quadruplex DNA (right) according to ICD analysis (gray: G4 quartet; red: ligand).
Figure 4: The simplified structure of the complex between 1e3 and quadruplex DNA (left; [38]) and the proposed or...
Unfortunately, the experimental data do not provide any evidence for a relevant effect of the adenine unit on the binding affinity of the ligands 4a–e or the complex structures with G4-DNA, because in this case significantly stronger differences of the binding constants, selectivities or optical responses should have been observed with the variation of the linker length. It may be concluded that the triazole ring, used as a synthetically convenient connection unit, imposes too much steric hindrance and restricted conformational flexibility in the vicinity of the adenine unit thus hindering the binding of the latter to the thymidine residues in the loops.
Conclusion
In summary, we have synthesized five novel berberine–adenine derivatives 4a–e with different lengths of the alkyltriazole linker units, which show the characteristic properties of berberine-based G4-DNA ligands. Notably, the binding affinities of the ligands do not change strongly with the length of the alkyl chain n and there is no obvious relationship between these parameters. Nevertheless, depending on the structure the ligands exhibit some significantly different effects when bound to the G4-DNA, such as fluorescent light-up effects and the formation of ICD bands, which are mostly pronounced with linker length of n = 4 (with a2) and n = 5 (with 22AG). This significant influence of the complex structure on the optical properties of the ligand provides some evidence that each ligand–G4-DNA complex has a specific structure with respect to the relative alignment and conformational flexibility of the ligand in the binding site. Considering these changes upon variation of either the ligand structure or the quadruplex form, the ligands 4a–e may have the potential to operate as selective ligands for G4-DNA, as indeed was shown exemplarily with the ligand 4e. The latter has a high selectivity to quadruplex DNA in competition with duplex DNA and stabilizes preferentially the G4-DNA forms 22AG and KRAS. We conclude from these results, along with the already reported data in the literature [36,37,39,42], that the derivatization of berberine by the attachment of functional substituents at the 9-position is a reasonable approach to fine-tune the binding properties with G4-DNA. However, more systematic investigations and broader structural variations are necessary to identify all relevant factors that affect the affinity and selectivity of such ligands.
Experimental
Equipment
NMR data were recorded with a Varian VNMR-S600 spectrometer [600 MHz (1H), 150 MHz (13C)] at 35 °C. NMR spectra were processed with the software ACD/NMR Processor Academic Version 12.01 and are referenced to the corresponding solvent [δ(DMSO-d5) = 2.50 (1H) and 39.5 (13C); δ(CHCl3) = 7.26 (1H) and 77.2 (13C)]. Elemental analyses data were determined with a HEKAtech EURO EA combustion analyzer by Mr. Rochus Breuer (Organic Chemistry I, University of Siegen). Mass spectra (ESI) were recorded on a Finnigan LCQ Deca (U = 6 kV; working gas: argon; auxiliary gas: nitrogen; temperature of the capillary: 200 °C). Absorption spectra were obtained with a Varian Cary 100 bio spectrometer in quartz cells (10 mm) with baseline correction. Emission spectra were recorded in quartz cells (10 mm) with a Cary Eclipse spectrometer at 20 °C. CD-spectroscopic measurements were performed with a Chirascan spectrometer (Applied Photophysics) in quartz cells (1.0 mm). Melting points were measured with a Büchi 545 (Büchi, Flawil, CH) and are uncorrected.
Materials
Commercially available reagents and reactants were used without further purification. Chemicals were obtained from the following companies: Carl Roth GmbH & Co. KG (Karlsruhe, D): Berberine chloride, 3-bromo-1-propyne. Alfa Aesar GmbH & Co. KG (Karlsruhe, D): Adenine. Acros Organics: 3-Phenyl-1-propyne. Biomers.net GmbH (Ulm, D): 22AG, a2, F21T, Fa2T, FmycT, FkitT, and FkrasT. 9-(Azidoalkyl)berberine derivatives 3a–e [54] and 9-propargyladenine (2) [53] were synthesized according to published procedures. Stock solutions of the ligands (1.0 mM) were prepared in MeOH (HPLC grade). Buffer solutions were prepared with biochemical grade chemicals in E-Pure water (resistivity ≥ 18 MΩ cm) and filtered through a membrane filter (0.45 µm pore size). K-phosphate-buffer (22AG, a2): 25 mM K2HPO4, 70 mM KCl; pH 7.0; Na-cacodylate buffer (F21T, Fa2T, FmycT, FkitT, and FkrasT): 90 µM LiCl, 10 mM KCl, 10 mM Na(CH3)2AsO2 × 3H2O, pH 7.2–7.3. The K-phosphate buffer solutions and the Na-cacodylate buffer solutions were stored at 4 °C in the dark.
Methods
The spectrophotometric and spectrofluorometric titrations with quadruplex DNA were performed according to published protocols [69]. To ensure a sufficient solubility during the titrations DMSO (1% v/v in BPE buffer and 5% v/v in K+-phosphate buffer) was used as a cosolvent.
For the CD spectra, solutions of G4-DNA in K+-phosphate butter and the ligands in buffer/DMSO were recorded after an equilibration time of 30 min.
For the thermal DNA denaturation analyses stock solutions of the ligand (20 µM) and the G4-DNA (50 µM) in Na-cacodylate buffer were used to prepare analyte solutions with different ligand:DNA ratios (LDR = 0.00, 1.25, 2.50, 5.00).
Synthesis
General procedure (GP) [54]
To a solution of the berberine azide derivatives 3a–e (1.0 molar equiv) and 9-propargyladenine (2, 1.1 molar equiv) in THF/MeCN 2:1 was added a solution of CuSO4 (0.3 equiv) and Na-ascorbate (1.1 molar equiv) in H2O. The mixture was stirred under reflux for 3 h. The solvent was removed under reduced pressure and the brown residue was dissolved in DMSO (15 mL) and filtered through a pad of neutral aluminum oxide. The pad was washed with DMSO (15 mL). The DMSO fractions were combined and the solvent was removed in vacuum. The remaining yellow solid was suspended in MeCN (500 mL) and filtered through a short pad of celite. The solvent was evaporated under reduced pressure and the crude product was purified by column chromatography (SiO2, CH2Cl2/MeOH 5→10%). After crystallization of the major fraction from MeOH/Et2O 7:3 the desired product was obtained as yellow solid.
9-O-{β-{4'-[(9''-Adenyl)methyl]-1H-1',2',3'-triazol-1'-yl}ethyl}berberine bromide (4a)
According to the GP, a solution of 3a (150 mg, 292 µmol), 2 (60.6 mg, 350 µmol), CuSO4 (5.83 mg, 36.5 µmol) and Na-ascorbate (36.3 mg, 183 µmol) was stirred at reflux in THF/H2O/MeCN 2:2:1 (25 mL). After purification by column chromatography (Rf = 0.11; 10% MeOH) and crystallization the product 4a was obtained as orange-colored amorphous solid (30.0 mg, 46.5 µmol, 16%); mp 191–194 °C (dec.); 1H NMR (600 MHz, DMSO-d6) δ 3.20 (t, 3J = 6.5 Hz, 2H, 5-H), 3.94 (s, 3H, O-CH3), 4.70 (t, 3J = 5.0 Hz, 2H,α-H), 4.84 (t, 3J = 6.5 Hz, 2H, 6-H), 4.90 (t, 3J = 5.0 Hz, 2H, β-H), 5.44 (s, 2H, C-CH2-N) 6.18 (s, 2H, O-CH2-O), 7.09 (s, 1H, 4-H), 7.23 (s, 2H, NH2) 7.77 (s, 1H, 1-H), 7.93 (d, 3J = 9.2 Hz, 12-H), 8.04 (s, 1H, 2''-H), 8.10 (d, 3J = 9.2 Hz, 1H, 11-H), 8.19 (s, 1H, 8''-H), 8.30 (s, 1H, 5'-H), 8.89 (s, 1H, 13-H), 9.48 (s, 1H, 8-H); 13C NMR (150 MHz, DMSO-d6) δ 26.3 (C5), 38.0 (C-CH2-N), 50.0 (Cβ), 55.4 (C6), 57.0 (O-CH3), 72.1 (Cα), 102.1 (O-CH2-O), 105.5 (C1), 108.4 (C4), 118.5 (C5''), 120.2 (C13), 120.3 (C1a), 121.2 (C8a), 123.8 (C12), 124.2 (C5'), 126.4 (C11), 130.6 (C4a), 132.8 (C12a), 137.4 (C13a), 140.7 (C8''), 141.7 (C9), 143.0 (C4'), 145.0 (C8), 147.7 (C2), 149.2 (C4''), 149.9 (C3), 150.0 (C10), 152.5 (C2''), 155.9 (C6''); ESIMS (m/z): 564 [M+]; anal. calcd for C29H26BrN9O4 (%): C, 54.05; H, 4.07; N, 19.56; found (%): C, 54.03; H, 4.41; N, 19.44.
9-O-{γ-{4'-[(9''-Adenyl)methyl]-1H-1',2',3'-triazol-1'-yl}propyl}berberine bromide (4b)
According to the GP, a solution of 3b (120 mg, 247 µmol), 2 (60.6 mg, 350 µmol), CuSO4 (5.01 mg, 31.4 µmol), and Na-ascorbate (47.1 mg, 272 µmol) was stirred at reflux in THF/H2O/MeCN 2:2:1 (25 mL). After purification by column chromatography (Rf = 0.12, 10% MeOH) and crystallization the product 4b was obtained as yellow, amorphous solid (42.3 mg, 64.0 µmol, 26%); mp 189–192 °C (dec.); 1H NMR (600 MHz, DMSO-d6) δ 2.41 (tt, 3J = 7 Hz, 3J = 7 Hz, 2H, β-H), 3.21 (t, 3J = 6 Hz, 2H, 5-H), 3.99 (s, 3H, O-CH3), 4.25 (t, 3J = 6 Hz, 2H, α-H), 4.67 (t, 3J = 7 Hz, 2H, γ-H), 4.94 (t, 3J = 6 Hz, 2H, 6-H), 5.45 (s, 2H, C-CH2-N), 6.18 (s, 2H, O-CH2-O), 7.09 (s, 1H, 4-H), 7.21 (s, 2H, NH2), 7.80 (s, 1H, 1-H), 7.99 (d, 3J = 9 Hz, 12-H), 8.08 (s, 1H, 2''-H), 8.16 (d, 3J = 9 Hz, 1H, 11-H), 8.21 (s, 1H, 8''-H), 8.24 (s, 1H, 5'-H), 8.94 (s, 1H, 13-H), 9.77 (s, 1H, 8-H); 13C NMR (150 MHz, DMSO-d6) δ 26.3 (5-C), 30.2 (Cβ), 38.1 (C-CH2-N), 46.6 (Cγ), 55.2 (C6), 57.0 (O-CH3), 71.7 (Cα), 102.1 (O-CH2-O), 105.5 (C1), 108.4 (C4), 118.5 (C5''), 120.2 (C13), 120.4 (C1a), 121.4 (C8a), 123.6 (C12), 123.8 (C5'), 126.6 (C11), 130.7 (C4a), 133.0 (C12a), 137.5 (C13a), 140.7 (C8''), 142.3 (C9), 142.7 (C4'), 145.2 (C8), 147.7 (C2), 149.3 (C4''), 149.8 (C3), 150.2 (C10), 152.5 (C2''), 155.9 (C6''); ESIMS (m/z): 578 [M+]; anal. calcd for C30H28BrN9O4 (%): C, 54.72; H, 4.29, N, 19.14; found (%): C, 54.69; H, 4.41; N, 18.79.
9-O-{δ-{4'-[(9''-Adenyl)methyl]-1H-1',2',3'-triazol-1'-yl}butyl}berberine bromide (4c)
According to the GP, a solution of 3c (240 mg, 481 µmol), 2 (96.1 mg, 529 µmol), CuSO4 (10.3 mg, 64.5 µmol), and Na-ascorbate (53.5 mg, 270 µmol) was stirred at reflux in THF/H2O/MeCN 2:2:1 (35 mL). After purification by column chromatography (Rf = 0.17, 10%) and crystallization the product 4c was obtained as yellow, amorphous solid (64.9 mg, 96.5 µmol, 20%); mp 157–158 °C (dec.); 1H NMR (600 MHz, DMSO-d6) δ 1.81 (tt, 3J = 7 Hz, 3J = 7 Hz, 2H, β-H), 2.06 (tt, 3J = 7 Hz, 3J = 7 Hz, 2H, γ-H), 3.21 (t, 3J = 6 Hz, 2H, 5-H), 4.02 (s, 3H, O-CH3), 4.26 (t, 3J = 6 Hz, 2H, α-H), 4.46 (t, 3J = 7 Hz, 2H, δ-H), 4.93 (t, 3J = 6 Hz, 2H, 6-H), 5.44 (s, 2H, C-CH2-N), 6.18 (s, 2H, O-CH2-O), 7.10 (s, 1H, 4-H), 7.23 (s, 2H, NH2), 7.80 (s, 1H, 1-H), 7.98 (d, 3J = 9 Hz, 12-H), 8.12 (s, 1H, 2''-H), 8.15 (s, 1H, 5'-H), 8.17 (d, 3J = 9 Hz, 1H, 11-H), 8.20 (s, 1H, 8''-H), 8.93 (s, 1H, 13-H), 9.78 (s, 1H, 8-H); 13C NMR (150 MHz, DMSO-d6) δ 26.3 (Cγ), 26.3 (C5), 26.5 (Cβ), 38.0 (C-CH2-N), 49.1 (Cδ), 55.3 (C6), 57.0 (O-CH3), 73.4 (Cα), 102.1 (O-CH2-O), 105.4 (C1), 108.4 (C4), 118.5 (C5''), 120.2 (C13), 120.4 (C1a), 121.5 (C8a), 123.4 (C12), 123.6 (C5'), 126.6 (C11), 130.7 (C4a), 133.0 (C12a), 137.5 (C13a), 140.6 (C8''), 142.5 (C4'), 142.6 (C9), 145.2 (C8), 147.7 (C2), 149.3 (C4''), 150.3 (C3), 152.5 (C2''), 155.9 (C6''); ESIMS (m/z): 592 [M+]; anal. calcd for C31H30BrN9O4 × H2O (%): C, 53.92; H, 4.67; N, 18.26; found (%): C, 54.07; H, 4.98; N, 18.07.
9-O-{ε-{4'-[(9''-Adenyl)methyl]-1H-1',2',3'-triazol-1'-yl}pentyl}berberine bromide (4d)
According to the GP, a solution of 3d (150 mg, 292 µmol), 2 (60.6 mg, 350 µmol), CuSO4 (5.83 mg, 36.5 µmol), and Na-ascorbate (36.6 mg, 183 µmol) was stirred at reflux in THF/H2O/MeCN 2:2:1 (25 mL). After purification by column chromatography (Rf = 0.19, 10% MeOH) and crystallization the product 4d was obtained as yellow, amorphous solid (73.2 mg, 107 µmol, 37%); mp 154–156 °C (dec.); 1H NMR (600 MHz, DMSO-d6) δ 1.41 (tt, 3J = 8 Hz, 3J = 7 Hz, 2H, γ-H), 1.86 (tt, 3J = 8 Hz, 3J = 7 Hz, 2H, β-H), 1.90 (tt, 3J = 7 Hz, 3J = 7 Hz, 2H, δ-H), 3.20 (t, 3J = 6 Hz, 2H, 5-H), 4.02 (s, 3H, O-CH3), 4.24 (t, 3J = 7 Hz, 2H, α-H), 4.39 (t, 3J = 7 Hz, 2H, ε-H), 4.95 (t, 3J = 6 Hz, 2H, 6-H), 5.42 (s, 2H, C-CH2-N), 6.17 (s, 2H, O-CH2-O), 7.08 (s, 1H, 4-H), 7.21 (s, 2H, NH2), 7.79 (s, 1H, 1-H), 7.98 (d, 3J = 9 Hz, 12-H), 8.11 (s, 1H, 2''-H), 8.14 (s, 1H, 5'-H), 8.16 (d, 3J = 9 Hz, 1H, 11-H), 8.19 (s, 1H, 8''-H), 8.94 (s, 1H, 13-H), 9.72 (s, 1H, 8-H); 13C NMR (150 MHz, DMSO-d6) δ 22.3 (Cγ), 26.3 (C5), 28.8 (Cβ), 29.4 (Cδ), 38.1 (C-CH2-N), 49.4 (Cε), 55.3 (C6), 57.0 (O-CH3), 73.9 (Cα), 102.1 (O-CH2-O), 105.5 (C1), 108.4 (C4), 118.6 (C5''), 120.3 (C13), 120.5 (C1a), 121.6 (C8a), 123.4 (C12), 123.5 (C5'), 126.6 (C11), 130.7 (C4a), 133.0 (C12a), 137.4 (C13a), 140.7 (C8''), 142.5 (C4'), 147.7 (C2), 149.3 (C4''), 149.8 (C3), 150.4 (C10), 152.5 (C2''), 155.9 (C6''); ESIMS (m/z): 606 [M+]; anal. calcd for C32H32BrN9O4 (%): C, 55.98; H, 4.70; N, 18.36; found (%): C, 55.70; H, 4.71; N, 18.11.
9-O-{ζ-{4'-[(9''-Adenyl)methyl]-1H-1',2',3'-triazol-1'-yl}hexyl}berberine bromide (4e)
According to the GP, a solution of 3e (300 mg, 567 µmol), 2 (118 mg, 682 µmol), CuSO4 (11.3 mg, 70.9 µmol), and Na-ascorbate (71.0 mg, 355 µmol) was stirred at reflux in THF/H2O/MeCN 2:2:1 (50 mL). After purification by column chromatography (Rf = 0.20, 10% MeOH) and crystallization the product 4e was obtained as yellow, amorphous solid (151 mg, 216 µmol, 38%); mp 152–154 °C (dec.); 1H NMR (600 MHz, DMSO-d6) δ 1.27–1.32 (m, 2H, δ-H), 1.45–1.49 (m, 2H, γ-H), 1.80–1.85 (m, 4H, β-H, ε-H), 3.20 (t, 3J = 6.2 Hz, 2H, 5-H), 4.02 (s, 3H, O-CH3), 4.23 (t, 3J = 6.8 Hz, 2H, α-H), 4.35 (t, 3J = 7.0 Hz, 2H, ζ-H), 4.97 (t, 3J = 6.2 Hz, 2H, 6-H), 5.43 (s, 2H, C-CH2-N), 6.17 (s, 2H, O-CH2-O), 7.07 (s, 1H, 4-H), 7.22 (s, 2H, NH2), 7.78 (s, 1H, 1-H), 7.98 (d, 3J = 9.2 Hz, 12-H), 8.12 (s, 1H, 2''-H), 8.15 (s, 1H, 5'-H), 8.16 (d, 3J = 9.2 Hz, 1H, 11-H), 8.20 (s, 1H, 8''-H), 8.97 (s, 1H, 13-H), 9.75 (s, 1H, 8-H); 13C NMR (150 MHz, DMSO-d6) δ 24.6 (Cγ), 25.5 (Cβ), 26.3 (C5), 29.2 (Cδ), 29.6 (Cε), 38.0 (C-CH2-N), 50.0 (Cζ), 55.4 (C6), 57.0 (O-CH3), 72.1 (Cα), 102.1 (O-CH2-O), 105.5 (C1), 108.5 (C4), 118.5 (C5''), 120.2 (C13), 120.4 (C1a), 121.6 (C8a), 123.3 (C12), 123.5 (C5'), 126.6 (C11), 130.6 (C4a), 133.0 (C12a), 137.4 (C13a), 140.6 (C8''), 142.8 (C4'), 147.7 (C2), 149.2 (C4''), 149.9 (C3), 150.3 (C10), 152.5 (C2''), 155.9 (C6''); ESIMS (m/z): 620 [M+]; anal. calcd for C32H34BrN9O4 × 3H2O (%): C, 52.52; H, 5.34; N, 16.70; found (%): C, 52.54; H, 5.00; N, 16.39.
Supporting Information
| Supporting Information File 1: Experimental procedures, additional spectroscopic data, 1H NMR and 13C NMR spectra. | ||
| Format: PDF | Size: 2.4 MB | Download |
References
-
Neidle, S. Nat. Rev. Chem. 2017, 1, 0041. doi:10.1038/s41570-017-0041
Return to citation in text: [1] [2] -
Neidle, S. J. Med. Chem. 2016, 59, 5987–6011. doi:10.1021/acs.jmedchem.5b01835
Return to citation in text: [1] [2] [3] -
Tateishi-Karimata, H.; Sugimoto, N. Chem. Commun. 2020, 56, 2379–2390. doi:10.1039/c9cc09771f
Return to citation in text: [1] -
Cañeque, T.; Müller, S.; Rodriguez, R. Nat. Rev. Chem. 2018, 2, 202–215. doi:10.1038/s41570-018-0030-x
Return to citation in text: [1] -
Laguerre, A.; Wong, J. M. Y.; Monchaud, D. Sci. Rep. 2016, 6, 32141. doi:10.1038/srep32141
Return to citation in text: [1] -
Chambers, V. S.; Marsico, G.; Boutell, J. M.; Di Antonio, M.; Smith, G. P.; Balasubramanian, S. Nat. Biotechnol. 2015, 33, 877–881. doi:10.1038/nbt.3295
Return to citation in text: [1] -
Verma, A.; Yadav, V. K.; Basundra, R.; Kumar, A.; Chowdhury, S. Nucleic Acids Res. 2009, 37, 4194–4204. doi:10.1093/nar/gkn1076
Return to citation in text: [1] -
Huppert, J. L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. Nucleic Acids Res. 2008, 36, 6260–6268. doi:10.1093/nar/gkn511
Return to citation in text: [1] -
Huppert, J. L.; Balasubramanian, S. Nucleic Acids Res. 2007, 35, 406–413. doi:10.1093/nar/gkl1057
Return to citation in text: [1] -
Collie, G. W.; Parkinson, G. N. Chem. Soc. Rev. 2011, 40, 5867–5892. doi:10.1039/c1cs15067g
Return to citation in text: [1] -
Li, Y.; Syed, J.; Suzuki, Y.; Asamitsu, S.; Shioda, N.; Wada, T.; Sugiyama, H. ChemBioChem 2016, 17, 928–935. doi:10.1002/cbic.201500655
Return to citation in text: [1] -
Balasubramanian, S.; Hurley, L. H.; Neidle, S. Nat. Rev. Drug Discovery 2011, 10, 261–275. doi:10.1038/nrd3428
Return to citation in text: [1] -
Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. doi:10.1038/nrm.2017.3
Return to citation in text: [1] -
Balasubramanian, S.; Neidle, S. Curr. Opin. Chem. Biol. 2009, 13, 345–353. doi:10.1016/j.cbpa.2009.04.637
Return to citation in text: [1] -
Monsen, R. C.; Trent, J. O. Biochimie 2018, 152, 134–148. doi:10.1016/j.biochi.2018.06.024
Return to citation in text: [1] -
Lee, H.-S.; Carmena, M.; Liskovykh, M.; Peat, E.; Kim, J.-H.; Oshimura, M.; Masumoto, H.; Teulade-Fichou, M.-P.; Pommier, Y.; Earnshaw, W. C.; Larionov, V.; Kouprina, N. Cancer Res. 2018, 78, 6282–6296. doi:10.1158/0008-5472.can-18-0894
Return to citation in text: [1] -
Nagatsugi, F.; Onizuka, K. Chem. Lett. 2020, 49, 771–780. doi:10.1246/cl.200214
Return to citation in text: [1] [2] [3] -
Asamitsu, S.; Bando, T.; Sugiyama, H. Chem. – Eur. J. 2019, 25, 417–430. doi:10.1002/chem.201802691
Return to citation in text: [1] [2] -
Dhamodharan, V.; Pradeepkumar, P. I. ACS Chem. Biol. 2019, 14, 2102–2114. doi:10.1021/acschembio.9b00475
Return to citation in text: [1] [2] -
O'Hagan, M. P.; Morales, J. C.; Galan, M. C. Eur. J. Org. Chem. 2019, 4995–5017. doi:10.1002/ejoc.201900692
Return to citation in text: [1] [2] -
Ruggiero, E.; Richter, S. N. Nucleic Acids Res. 2018, 46, 3270–3283. doi:10.1093/nar/gky187
Return to citation in text: [1] [2] -
Duarte, A. R.; Cadoni, E.; Ressurreição, A. S.; Moreira, R.; Paulo, A. ChemMedChem 2018, 13, 869–893. doi:10.1002/cmdc.201700747
Return to citation in text: [1] [2] -
Gao, Y.; Wang, F.; Song, Y.; Liu, H. Chin. Med. (London, U. K.) 2020, 15, 7. doi:10.1186/s13020-020-0288-z
Return to citation in text: [1] -
Chu, M.; Chen, X.; Wang, J.; Guo, L.; Wang, Q.; Gao, Z.; Kang, J.; Zhang, M.; Feng, J.; Guo, Q.; Li, B.; Zhang, C.; Guo, X.; Chu, Z.; Wang, Y. Front. Pharmacol. 2018, 9, 801. doi:10.3389/fphar.2018.00801
Return to citation in text: [1] -
Kim, D.-G.; Choi, J.-W.; Jo, I.-J.; Kim, M.-J.; Lee, H.-S.; Hong, S.-H.; Song, H.-J.; Bae, G.-S.; Park, H.-S. Mol. Med. Rep. 2020, 21, 258–266. doi:10.3892/mmr.2019.10823
Return to citation in text: [1] -
Li, Q.; He, W.; Zhang, L.; Zu, Y.; Zhu, Q.; Deng, X.; Zhao, T.; Gao, W.; Zhang, B. Lett. Drug Des. Discovery 2012, 9, 573–580. doi:10.2174/157018012800673010
Return to citation in text: [1] -
Yi, Z.-B.; Yan, Y.; Liang, Y.-Z.; Bao, Z. J. Pharm. Biomed. Anal. 2007, 44, 301–304. doi:10.1016/j.jpba.2007.02.018
Return to citation in text: [1] -
Jin, F.; Xie, T.; Huang, X.; Zhao, X. Pharm. Biol. (Abingdon, U. K.) 2018, 56, 665–671. doi:10.1080/13880209.2018.1548627
Return to citation in text: [1] -
Wang, Y.; Zhang, S. Biomed. Pharmacother. 2018, 103, 1287–1293. doi:10.1016/j.biopha.2018.04.161
Return to citation in text: [1] -
Vieira, S.; Castelli, S.; Falconi, M.; Takarada, J.; Fiorillo, G.; Buzzetti, F.; Lombardi, P.; Desideri, A. Int. J. Biol. Macromol. 2015, 77, 68–75. doi:10.1016/j.ijbiomac.2015.02.051
Return to citation in text: [1] -
Chu, S.-C.; Yu, C.-C.; Hsu, L.-S.; Chen, K.-S.; Su, M.-Y.; Chen, P.-N. Mol. Pharmacol. 2014, 86, 609–623. doi:10.1124/mol.114.094037
Return to citation in text: [1] -
Lee, K.-H.; Lo, H.-L.; Tang, W.-C.; Hsiao, H. H.-y.; Yang, P.-M. Sci. Rep. 2014, 4, 6394. doi:10.1038/srep06394
Return to citation in text: [1] -
Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2136–E2145. doi:10.1073/pnas.1612627114
Return to citation in text: [1] [2] [3] -
Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A. Y.; Kumar, G. S.; Gratteri, P. ACS Med. Chem. Lett. 2020, 11, 645–650. doi:10.1021/acsmedchemlett.9b00516
Return to citation in text: [1] [2] [3] [4] -
Xiong, Y.-X.; Su, H.-F.; Lv, P.; Ma, Y.; Wang, S.-K.; Miao, H.; Liu, H.-Y.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S. Oncotarget 2015, 6, 35625–35635. doi:10.18632/oncotarget.5521
Return to citation in text: [1] [2] [3] -
Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029
Return to citation in text: [1] [2] [3] [4] [5] -
Zhang, W.-J.; Ou, T.-M.; Lu, Y.-J.; Huang, Y.-Y.; Wu, W.-B.; Huang, Z.-S.; Zhou, J.-L.; Wong, K.-Y.; Gu, L.-Q. Bioorg. Med. Chem. 2007, 15, 5493–5501. doi:10.1016/j.bmc.2007.05.050
Return to citation in text: [1] [2] [3] [4] -
Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] [8] -
Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486
Return to citation in text: [1] [2] [3] [4] [5] -
Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a
Return to citation in text: [1] [2] [3] [4] [5] -
Islam, M. M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Suresh Kumar, G. DNA Cell Biol. 2011, 30, 123–133. doi:10.1089/dna.2010.1109
Return to citation in text: [1] [2] [3] [4] -
Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030
Return to citation in text: [1] [2] [3] [4] [5] -
Wang, M.-Q.; Xu, J.; Zhang, L.; Liao, Y.; Wei, H.; Yin, Y.-Y.; Liu, Q.; Zhang, Y. Bioorg. Med. Chem. 2019, 27, 552–559. doi:10.1016/j.bmc.2018.12.037
Return to citation in text: [1] -
Funke, A.; Weisz, K. Biochimie 2019, 157, 142–148. doi:10.1016/j.biochi.2018.11.015
Return to citation in text: [1] -
Lavrado, J.; Ohnmacht, S. A.; Correia, I.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Moreira, R.; Neidle, S.; dos Santos, D. J. V. A.; Paulo, A. ChemMedChem 2015, 10, 836–849. doi:10.1002/cmdc.201500067
Return to citation in text: [1] -
Singh, M.; Wang, S.; Joo, H.; Ye, Z.; Christison, K. M.; Hekman, R.; Vierra, C.; Xue, L. Chem. Biol. Drug Des. 2020. doi:10.1111/cbdd.13741
Return to citation in text: [1] -
Tsvetkov, V. B.; Varizhuk, A. M.; Lizunova, S. A.; Nikolenko, T. A.; Ivanov, I. A.; Severov, V. V.; Belyaev, E. S.; Shitikov, E. A.; Pozmogova, G. E.; Aralov, A. V. Org. Biomol. Chem. 2020, 18, 6147–6154. doi:10.1039/d0ob00983k
Return to citation in text: [1] -
Wang, S.; Yang, D.; Singh, M.; Joo, H.; Rangel, V. M.; Tran, A.; Phan, E.; Xue, L. Eur. J. Med. Chem. 2019, 175, 20–33. doi:10.1016/j.ejmech.2019.04.041
Return to citation in text: [1] -
Zhang, L.; Er, J. C.; Li, X.; Heng, J. J.; Samanta, A.; Chang, Y.-T.; Lee, C.-L. K. Chem. Commun. 2015, 51, 7386–7389. doi:10.1039/c5cc01601k
Return to citation in text: [1] [2] -
Yu, Z.; Schonhoft, J. D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. J. Am. Chem. Soc. 2009, 131, 1876–1882. doi:10.1021/ja806782s
Return to citation in text: [1] [2] -
Dzubiel, D.; Ihmels, H.; Mahmoud, M. M. A.; Thomas, L. Beilstein J. Org. Chem. 2014, 10, 2963–2974. doi:10.3762/bjoc.10.314
Return to citation in text: [1] [2] -
Saha, P.; Panda, D.; Dash, J. Chem. Commun. 2019, 55, 731–750. doi:10.1039/c8cc07107a
Return to citation in text: [1] -
Wu, Y.-C.; Kuo, S.-W. J. Mater. Chem. 2012, 22, 2982–2991. doi:10.1039/c1jm14699h
Return to citation in text: [1] [2] -
Halimani, M.; Chandran, S. P.; Kashyap, S.; Jadhav, V. M.; Prasad, B. L. V.; Hotha, S.; Maiti, S. Langmuir 2009, 25, 2339–2347. doi:10.1021/la802761b
Return to citation in text: [1] [2] [3] -
Megyesi, M.; Biczók, L.; Jablonkai, I. J. Phys. Chem. C 2008, 112, 3410–3416. doi:10.1021/jp075348w
Return to citation in text: [1] -
Díaz, M. S.; Freile, M. L.; Gutiérrez, M. I. Photochem. Photobiol. Sci. 2009, 8, 970–974. doi:10.1039/b822363g
Return to citation in text: [1] -
Huang, X.; Zhang, X.; Qian, T.; Ma, J.; Cui, L.; Li, C. Beilstein J. Org. Chem. 2018, 14, 2236–2241. doi:10.3762/bjoc.14.198
Return to citation in text: [1] -
Colina, A. N.; Díaz, M. S.; Gutiérrez, M. I. J. Lumin. 2013, 144, 198–202. doi:10.1016/j.jlumin.2013.07.023
Return to citation in text: [1] -
Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Methods 2012, 57, 64–75. doi:10.1016/j.ymeth.2012.03.011
Return to citation in text: [1] [2] -
Rezler, E. M.; Seenisamy, J.; Bashyam, S.; Kim, M.-Y.; White, E.; Wilson, W. D.; Hurley, L. H. J. Am. Chem. Soc. 2005, 127, 9439–9447. doi:10.1021/ja0505088
Return to citation in text: [1] -
Timmer, C. M.; Michmerhuizen, N. L.; Witte, A. B.; Van Winkle, M.; Zhou, D.; Sinniah, K. J. Phys. Chem. B 2014, 118, 1784–1790. doi:10.1021/jp411293r
Return to citation in text: [1] -
Schonhoft, J. D.; Das, A.; Achamyeleh, F.; Samdani, S.; Sewell, A.; Mao, H.; Basu, S. Biopolymers 2010, 93, 21–31. doi:10.1002/bip.21289
Return to citation in text: [1] -
Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Beilstein J. Org. Chem. 2018, 14, 84–105. doi:10.3762/bjoc.14.5
Return to citation in text: [1] [2] -
Norden, B.; Rodger, A.; Dafforn, T. Linear Dichroism and Circular Dichroism; RSC Publishing: Cambridge, UK, 2010.
Return to citation in text: [1] [2] -
Nordén, B.; Kurucsev, T. J. Mol. Recognit. 1994, 7, 141–155. doi:10.1002/jmr.300070211
Return to citation in text: [1] -
Zhou, C.-Q.; Yang, J.-W.; Dong, C.; Wang, Y.-M.; Sun, B.; Chen, J.-X.; Xu, Y.-S.; Chen, W.-H. Org. Biomol. Chem. 2016, 14, 191–197. doi:10.1039/c5ob01723h
Return to citation in text: [1] -
Li, Z.-Q.; Liao, T.-C.; Dong, C.; Yang, J.-W.; Chen, X.-J.; Liu, L.; Luo, Y.; Liang, Y.-Y.; Chen, W.-H.; Zhou, C.-Q. Org. Biomol. Chem. 2017, 15, 10221–10229. doi:10.1039/c7ob02326j
Return to citation in text: [1] -
Tera, M.; Hirokawa, T.; Okabe, S.; Sugahara, K.; Seimiya, H.; Shimamoto, K. Chem. – Eur. J. 2015, 21, 14519–14528. doi:10.1002/chem.201501693
Return to citation in text: [1] -
Bortolozzi, R.; Ihmels, H.; Thomas, L.; Tian, M.; Viola, G. Chem. – Eur. J. 2013, 19, 8736–8741. doi:10.1002/chem.201301164
Return to citation in text: [1]
| 52. | Saha, P.; Panda, D.; Dash, J. Chem. Commun. 2019, 55, 731–750. doi:10.1039/c8cc07107a |
| 53. | Wu, Y.-C.; Kuo, S.-W. J. Mater. Chem. 2012, 22, 2982–2991. doi:10.1039/c1jm14699h |
| 54. | Halimani, M.; Chandran, S. P.; Kashyap, S.; Jadhav, V. M.; Prasad, B. L. V.; Hotha, S.; Maiti, S. Langmuir 2009, 25, 2339–2347. doi:10.1021/la802761b |
| 49. | Zhang, L.; Er, J. C.; Li, X.; Heng, J. J.; Samanta, A.; Chang, Y.-T.; Lee, C.-L. K. Chem. Commun. 2015, 51, 7386–7389. doi:10.1039/c5cc01601k |
| 59. | Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Methods 2012, 57, 64–75. doi:10.1016/j.ymeth.2012.03.011 |
| 55. | Megyesi, M.; Biczók, L.; Jablonkai, I. J. Phys. Chem. C 2008, 112, 3410–3416. doi:10.1021/jp075348w |
| 56. | Díaz, M. S.; Freile, M. L.; Gutiérrez, M. I. Photochem. Photobiol. Sci. 2009, 8, 970–974. doi:10.1039/b822363g |
| 57. | Huang, X.; Zhang, X.; Qian, T.; Ma, J.; Cui, L.; Li, C. Beilstein J. Org. Chem. 2018, 14, 2236–2241. doi:10.3762/bjoc.14.198 |
| 58. | Colina, A. N.; Díaz, M. S.; Gutiérrez, M. I. J. Lumin. 2013, 144, 198–202. doi:10.1016/j.jlumin.2013.07.023 |
| 50. | Yu, Z.; Schonhoft, J. D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. J. Am. Chem. Soc. 2009, 131, 1876–1882. doi:10.1021/ja806782s |
| 51. | Dzubiel, D.; Ihmels, H.; Mahmoud, M. M. A.; Thomas, L. Beilstein J. Org. Chem. 2014, 10, 2963–2974. doi:10.3762/bjoc.10.314 |
| 33. | Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2136–E2145. doi:10.1073/pnas.1612627114 |
| 34. | Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A. Y.; Kumar, G. S.; Gratteri, P. ACS Med. Chem. Lett. 2020, 11, 645–650. doi:10.1021/acsmedchemlett.9b00516 |
| 35. | Xiong, Y.-X.; Su, H.-F.; Lv, P.; Ma, Y.; Wang, S.-K.; Miao, H.; Liu, H.-Y.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S. Oncotarget 2015, 6, 35625–35635. doi:10.18632/oncotarget.5521 |
| 36. | Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029 |
| 37. | Zhang, W.-J.; Ou, T.-M.; Lu, Y.-J.; Huang, Y.-Y.; Wu, W.-B.; Huang, Z.-S.; Zhou, J.-L.; Wong, K.-Y.; Gu, L.-Q. Bioorg. Med. Chem. 2007, 15, 5493–5501. doi:10.1016/j.bmc.2007.05.050 |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 39. | Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486 |
| 40. | Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a |
| 41. | Islam, M. M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Suresh Kumar, G. DNA Cell Biol. 2011, 30, 123–133. doi:10.1089/dna.2010.1109 |
| 42. | Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030 |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 39. | Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486 |
| 40. | Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a |
| 41. | Islam, M. M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Suresh Kumar, G. DNA Cell Biol. 2011, 30, 123–133. doi:10.1089/dna.2010.1109 |
| 42. | Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030 |
| 17. | Nagatsugi, F.; Onizuka, K. Chem. Lett. 2020, 49, 771–780. doi:10.1246/cl.200214 |
| 18. | Asamitsu, S.; Bando, T.; Sugiyama, H. Chem. – Eur. J. 2019, 25, 417–430. doi:10.1002/chem.201802691 |
| 19. | Dhamodharan, V.; Pradeepkumar, P. I. ACS Chem. Biol. 2019, 14, 2102–2114. doi:10.1021/acschembio.9b00475 |
| 20. | O'Hagan, M. P.; Morales, J. C.; Galan, M. C. Eur. J. Org. Chem. 2019, 4995–5017. doi:10.1002/ejoc.201900692 |
| 21. | Ruggiero, E.; Richter, S. N. Nucleic Acids Res. 2018, 46, 3270–3283. doi:10.1093/nar/gky187 |
| 22. | Duarte, A. R.; Cadoni, E.; Ressurreição, A. S.; Moreira, R.; Paulo, A. ChemMedChem 2018, 13, 869–893. doi:10.1002/cmdc.201700747 |
| 33. | Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2136–E2145. doi:10.1073/pnas.1612627114 |
| 34. | Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A. Y.; Kumar, G. S.; Gratteri, P. ACS Med. Chem. Lett. 2020, 11, 645–650. doi:10.1021/acsmedchemlett.9b00516 |
| 35. | Xiong, Y.-X.; Su, H.-F.; Lv, P.; Ma, Y.; Wang, S.-K.; Miao, H.; Liu, H.-Y.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S. Oncotarget 2015, 6, 35625–35635. doi:10.18632/oncotarget.5521 |
| 36. | Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029 |
| 37. | Zhang, W.-J.; Ou, T.-M.; Lu, Y.-J.; Huang, Y.-Y.; Wu, W.-B.; Huang, Z.-S.; Zhou, J.-L.; Wong, K.-Y.; Gu, L.-Q. Bioorg. Med. Chem. 2007, 15, 5493–5501. doi:10.1016/j.bmc.2007.05.050 |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 39. | Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486 |
| 40. | Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a |
| 41. | Islam, M. M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Suresh Kumar, G. DNA Cell Biol. 2011, 30, 123–133. doi:10.1089/dna.2010.1109 |
| 42. | Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030 |
| 59. | Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Methods 2012, 57, 64–75. doi:10.1016/j.ymeth.2012.03.011 |
| 60. | Rezler, E. M.; Seenisamy, J.; Bashyam, S.; Kim, M.-Y.; White, E.; Wilson, W. D.; Hurley, L. H. J. Am. Chem. Soc. 2005, 127, 9439–9447. doi:10.1021/ja0505088 |
| 61. | Timmer, C. M.; Michmerhuizen, N. L.; Witte, A. B.; Van Winkle, M.; Zhou, D.; Sinniah, K. J. Phys. Chem. B 2014, 118, 1784–1790. doi:10.1021/jp411293r |
| 62. | Schonhoft, J. D.; Das, A.; Achamyeleh, F.; Samdani, S.; Sewell, A.; Mao, H.; Basu, S. Biopolymers 2010, 93, 21–31. doi:10.1002/bip.21289 |
| 64. | Norden, B.; Rodger, A.; Dafforn, T. Linear Dichroism and Circular Dichroism; RSC Publishing: Cambridge, UK, 2010. |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 68. | Tera, M.; Hirokawa, T.; Okabe, S.; Sugahara, K.; Seimiya, H.; Shimamoto, K. Chem. – Eur. J. 2015, 21, 14519–14528. doi:10.1002/chem.201501693 |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 34. | Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A. Y.; Kumar, G. S.; Gratteri, P. ACS Med. Chem. Lett. 2020, 11, 645–650. doi:10.1021/acsmedchemlett.9b00516 |
| 36. | Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029 |
| 40. | Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a |
| 66. | Zhou, C.-Q.; Yang, J.-W.; Dong, C.; Wang, Y.-M.; Sun, B.; Chen, J.-X.; Xu, Y.-S.; Chen, W.-H. Org. Biomol. Chem. 2016, 14, 191–197. doi:10.1039/c5ob01723h |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 67. | Li, Z.-Q.; Liao, T.-C.; Dong, C.; Yang, J.-W.; Chen, X.-J.; Liu, L.; Luo, Y.; Liang, Y.-Y.; Chen, W.-H.; Zhou, C.-Q. Org. Biomol. Chem. 2017, 15, 10221–10229. doi:10.1039/c7ob02326j |
| 63. | Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Beilstein J. Org. Chem. 2018, 14, 84–105. doi:10.3762/bjoc.14.5 |
| 64. | Norden, B.; Rodger, A.; Dafforn, T. Linear Dichroism and Circular Dichroism; RSC Publishing: Cambridge, UK, 2010. |
| 65. | Nordén, B.; Kurucsev, T. J. Mol. Recognit. 1994, 7, 141–155. doi:10.1002/jmr.300070211 |
| 63. | Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Beilstein J. Org. Chem. 2018, 14, 84–105. doi:10.3762/bjoc.14.5 |
| 54. | Halimani, M.; Chandran, S. P.; Kashyap, S.; Jadhav, V. M.; Prasad, B. L. V.; Hotha, S.; Maiti, S. Langmuir 2009, 25, 2339–2347. doi:10.1021/la802761b |
| 53. | Wu, Y.-C.; Kuo, S.-W. J. Mater. Chem. 2012, 22, 2982–2991. doi:10.1039/c1jm14699h |
| 36. | Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029 |
| 37. | Zhang, W.-J.; Ou, T.-M.; Lu, Y.-J.; Huang, Y.-Y.; Wu, W.-B.; Huang, Z.-S.; Zhou, J.-L.; Wong, K.-Y.; Gu, L.-Q. Bioorg. Med. Chem. 2007, 15, 5493–5501. doi:10.1016/j.bmc.2007.05.050 |
| 39. | Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486 |
| 42. | Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030 |
| 1. | Neidle, S. Nat. Rev. Chem. 2017, 1, 0041. doi:10.1038/s41570-017-0041 |
| 2. | Neidle, S. J. Med. Chem. 2016, 59, 5987–6011. doi:10.1021/acs.jmedchem.5b01835 |
| 3. | Tateishi-Karimata, H.; Sugimoto, N. Chem. Commun. 2020, 56, 2379–2390. doi:10.1039/c9cc09771f |
| 11. | Li, Y.; Syed, J.; Suzuki, Y.; Asamitsu, S.; Shioda, N.; Wada, T.; Sugiyama, H. ChemBioChem 2016, 17, 928–935. doi:10.1002/cbic.201500655 |
| 12. | Balasubramanian, S.; Hurley, L. H.; Neidle, S. Nat. Rev. Drug Discovery 2011, 10, 261–275. doi:10.1038/nrd3428 |
| 31. | Chu, S.-C.; Yu, C.-C.; Hsu, L.-S.; Chen, K.-S.; Su, M.-Y.; Chen, P.-N. Mol. Pharmacol. 2014, 86, 609–623. doi:10.1124/mol.114.094037 |
| 32. | Lee, K.-H.; Lo, H.-L.; Tang, W.-C.; Hsiao, H. H.-y.; Yang, P.-M. Sci. Rep. 2014, 4, 6394. doi:10.1038/srep06394 |
| 10. | Collie, G. W.; Parkinson, G. N. Chem. Soc. Rev. 2011, 40, 5867–5892. doi:10.1039/c1cs15067g |
| 33. | Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2136–E2145. doi:10.1073/pnas.1612627114 |
| 7. | Verma, A.; Yadav, V. K.; Basundra, R.; Kumar, A.; Chowdhury, S. Nucleic Acids Res. 2009, 37, 4194–4204. doi:10.1093/nar/gkn1076 |
| 8. | Huppert, J. L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. Nucleic Acids Res. 2008, 36, 6260–6268. doi:10.1093/nar/gkn511 |
| 9. | Huppert, J. L.; Balasubramanian, S. Nucleic Acids Res. 2007, 35, 406–413. doi:10.1093/nar/gkl1057 |
| 28. | Jin, F.; Xie, T.; Huang, X.; Zhao, X. Pharm. Biol. (Abingdon, U. K.) 2018, 56, 665–671. doi:10.1080/13880209.2018.1548627 |
| 29. | Wang, Y.; Zhang, S. Biomed. Pharmacother. 2018, 103, 1287–1293. doi:10.1016/j.biopha.2018.04.161 |
| 4. | Cañeque, T.; Müller, S.; Rodriguez, R. Nat. Rev. Chem. 2018, 2, 202–215. doi:10.1038/s41570-018-0030-x |
| 5. | Laguerre, A.; Wong, J. M. Y.; Monchaud, D. Sci. Rep. 2016, 6, 32141. doi:10.1038/srep32141 |
| 6. | Chambers, V. S.; Marsico, G.; Boutell, J. M.; Di Antonio, M.; Smith, G. P.; Balasubramanian, S. Nat. Biotechnol. 2015, 33, 877–881. doi:10.1038/nbt.3295 |
| 2. | Neidle, S. J. Med. Chem. 2016, 59, 5987–6011. doi:10.1021/acs.jmedchem.5b01835 |
| 30. | Vieira, S.; Castelli, S.; Falconi, M.; Takarada, J.; Fiorillo, G.; Buzzetti, F.; Lombardi, P.; Desideri, A. Int. J. Biol. Macromol. 2015, 77, 68–75. doi:10.1016/j.ijbiomac.2015.02.051 |
| 17. | Nagatsugi, F.; Onizuka, K. Chem. Lett. 2020, 49, 771–780. doi:10.1246/cl.200214 |
| 25. | Kim, D.-G.; Choi, J.-W.; Jo, I.-J.; Kim, M.-J.; Lee, H.-S.; Hong, S.-H.; Song, H.-J.; Bae, G.-S.; Park, H.-S. Mol. Med. Rep. 2020, 21, 258–266. doi:10.3892/mmr.2019.10823 |
| 17. | Nagatsugi, F.; Onizuka, K. Chem. Lett. 2020, 49, 771–780. doi:10.1246/cl.200214 |
| 18. | Asamitsu, S.; Bando, T.; Sugiyama, H. Chem. – Eur. J. 2019, 25, 417–430. doi:10.1002/chem.201802691 |
| 19. | Dhamodharan, V.; Pradeepkumar, P. I. ACS Chem. Biol. 2019, 14, 2102–2114. doi:10.1021/acschembio.9b00475 |
| 20. | O'Hagan, M. P.; Morales, J. C.; Galan, M. C. Eur. J. Org. Chem. 2019, 4995–5017. doi:10.1002/ejoc.201900692 |
| 21. | Ruggiero, E.; Richter, S. N. Nucleic Acids Res. 2018, 46, 3270–3283. doi:10.1093/nar/gky187 |
| 22. | Duarte, A. R.; Cadoni, E.; Ressurreição, A. S.; Moreira, R.; Paulo, A. ChemMedChem 2018, 13, 869–893. doi:10.1002/cmdc.201700747 |
| 26. | Li, Q.; He, W.; Zhang, L.; Zu, Y.; Zhu, Q.; Deng, X.; Zhao, T.; Gao, W.; Zhang, B. Lett. Drug Des. Discovery 2012, 9, 573–580. doi:10.2174/157018012800673010 |
| 27. | Yi, Z.-B.; Yan, Y.; Liang, Y.-Z.; Bao, Z. J. Pharm. Biomed. Anal. 2007, 44, 301–304. doi:10.1016/j.jpba.2007.02.018 |
| 1. | Neidle, S. Nat. Rev. Chem. 2017, 1, 0041. doi:10.1038/s41570-017-0041 |
| 2. | Neidle, S. J. Med. Chem. 2016, 59, 5987–6011. doi:10.1021/acs.jmedchem.5b01835 |
| 15. | Monsen, R. C.; Trent, J. O. Biochimie 2018, 152, 134–148. doi:10.1016/j.biochi.2018.06.024 |
| 16. | Lee, H.-S.; Carmena, M.; Liskovykh, M.; Peat, E.; Kim, J.-H.; Oshimura, M.; Masumoto, H.; Teulade-Fichou, M.-P.; Pommier, Y.; Earnshaw, W. C.; Larionov, V.; Kouprina, N. Cancer Res. 2018, 78, 6282–6296. doi:10.1158/0008-5472.can-18-0894 |
| 69. | Bortolozzi, R.; Ihmels, H.; Thomas, L.; Tian, M.; Viola, G. Chem. – Eur. J. 2013, 19, 8736–8741. doi:10.1002/chem.201301164 |
| 13. | Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. doi:10.1038/nrm.2017.3 |
| 14. | Balasubramanian, S.; Neidle, S. Curr. Opin. Chem. Biol. 2009, 13, 345–353. doi:10.1016/j.cbpa.2009.04.637 |
| 23. | Gao, Y.; Wang, F.; Song, Y.; Liu, H. Chin. Med. (London, U. K.) 2020, 15, 7. doi:10.1186/s13020-020-0288-z |
| 24. | Chu, M.; Chen, X.; Wang, J.; Guo, L.; Wang, Q.; Gao, Z.; Kang, J.; Zhang, M.; Feng, J.; Guo, Q.; Li, B.; Zhang, C.; Guo, X.; Chu, Z.; Wang, Y. Front. Pharmacol. 2018, 9, 801. doi:10.3389/fphar.2018.00801 |
| 54. | Halimani, M.; Chandran, S. P.; Kashyap, S.; Jadhav, V. M.; Prasad, B. L. V.; Hotha, S.; Maiti, S. Langmuir 2009, 25, 2339–2347. doi:10.1021/la802761b |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 34. | Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A. Y.; Kumar, G. S.; Gratteri, P. ACS Med. Chem. Lett. 2020, 11, 645–650. doi:10.1021/acsmedchemlett.9b00516 |
| 35. | Xiong, Y.-X.; Su, H.-F.; Lv, P.; Ma, Y.; Wang, S.-K.; Miao, H.; Liu, H.-Y.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S. Oncotarget 2015, 6, 35625–35635. doi:10.18632/oncotarget.5521 |
| 36. | Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. 2008, 16, 7582–7591. doi:10.1016/j.bmc.2008.07.029 |
| 37. | Zhang, W.-J.; Ou, T.-M.; Lu, Y.-J.; Huang, Y.-Y.; Wu, W.-B.; Huang, Z.-S.; Zhou, J.-L.; Wong, K.-Y.; Gu, L.-Q. Bioorg. Med. Chem. 2007, 15, 5493–5501. doi:10.1016/j.bmc.2007.05.050 |
| 38. | Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L. M.; Nocentini, A.; Scovassi, A. I.; Lombardi, P.; Gratteri, P. Chem. – Asian J. 2016, 11, 1107–1115. doi:10.1002/asia.201600116 |
| 39. | Bhowmik, D.; Fiorillo, G.; Lombardi, P.; Suresh Kumar, G. J. Mol. Recognit. 2015, 28, 722–730. doi:10.1002/jmr.2486 |
| 40. | Bhowmik, D.; Hossain, M.; Buzzetti, F.; D’Auria, R.; Lombardi, P.; Kumar, G. S. J. Phys. Chem. B 2012, 116, 2314–2324. doi:10.1021/jp210072a |
| 41. | Islam, M. M.; Basu, A.; Hossain, M.; Sureshkumar, G.; Hotha, S.; Suresh Kumar, G. DNA Cell Biol. 2011, 30, 123–133. doi:10.1089/dna.2010.1109 |
| 42. | Ma, Y.; Ou, T.-M.; Tan, J.-H.; Hou, J.-Q.; Huang, S.-L.; Gu, L.-Q.; Huang, Z.-S. Bioorg. Med. Chem. Lett. 2009, 19, 3414–3417. doi:10.1016/j.bmcl.2009.05.030 |
| 50. | Yu, Z.; Schonhoft, J. D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. J. Am. Chem. Soc. 2009, 131, 1876–1882. doi:10.1021/ja806782s |
| 51. | Dzubiel, D.; Ihmels, H.; Mahmoud, M. M. A.; Thomas, L. Beilstein J. Org. Chem. 2014, 10, 2963–2974. doi:10.3762/bjoc.10.314 |
| 48. | Wang, S.; Yang, D.; Singh, M.; Joo, H.; Rangel, V. M.; Tran, A.; Phan, E.; Xue, L. Eur. J. Med. Chem. 2019, 175, 20–33. doi:10.1016/j.ejmech.2019.04.041 |
| 49. | Zhang, L.; Er, J. C.; Li, X.; Heng, J. J.; Samanta, A.; Chang, Y.-T.; Lee, C.-L. K. Chem. Commun. 2015, 51, 7386–7389. doi:10.1039/c5cc01601k |
| 46. | Singh, M.; Wang, S.; Joo, H.; Ye, Z.; Christison, K. M.; Hekman, R.; Vierra, C.; Xue, L. Chem. Biol. Drug Des. 2020. doi:10.1111/cbdd.13741 |
| 47. | Tsvetkov, V. B.; Varizhuk, A. M.; Lizunova, S. A.; Nikolenko, T. A.; Ivanov, I. A.; Severov, V. V.; Belyaev, E. S.; Shitikov, E. A.; Pozmogova, G. E.; Aralov, A. V. Org. Biomol. Chem. 2020, 18, 6147–6154. doi:10.1039/d0ob00983k |
| 43. | Wang, M.-Q.; Xu, J.; Zhang, L.; Liao, Y.; Wei, H.; Yin, Y.-Y.; Liu, Q.; Zhang, Y. Bioorg. Med. Chem. 2019, 27, 552–559. doi:10.1016/j.bmc.2018.12.037 |
| 44. | Funke, A.; Weisz, K. Biochimie 2019, 157, 142–148. doi:10.1016/j.biochi.2018.11.015 |
| 45. | Lavrado, J.; Ohnmacht, S. A.; Correia, I.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Moreira, R.; Neidle, S.; dos Santos, D. J. V. A.; Paulo, A. ChemMedChem 2015, 10, 836–849. doi:10.1002/cmdc.201500067 |
© 2020 Becher et al.; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0). Please note that the reuse, redistribution and reproduction in particular requires that the authors and source are credited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (https://www.beilstein-journals.org/bjoc)