Abstract
A simple and straightforward synthetic approach for carbon nanodots (C-dots) is proposed. The strategy is based on a one-step hydrothermal chemical reduction with thiourea and urea, leading to high quantum yield C-dots. The obtained C-dots are well-dispersed with a uniform size and a graphite-like structure. A synergistic reduction mechanism was investigated using Fourier transform infrared spectroscopy and X-ray photoelectron spectroscopy. The findings show that using both thiourea and urea during the one-pot synthesis enhances the luminescence of the generated C-dots. Moreover, the prepared C-dots have a high distribution of functional groups on their surface. In this work, C-dots proved to be a suitable nanomaterial for imaging of bacteria and exhibit potential for application in bioimaging thanks to their low cytotoxicity.
Introduction
Over recent years, carbon nanomaterials have remarkably influenced the growth of a wide range of fields, including electronics, photonics, energy, catalysis and medicine. Within this class of materials, carbon nanodots (C-dots) are deemed a major breakthrough for the development of fluorescent nanomaterials. They are a promising alternative to fluorescent inorganic semiconductor nanocrystals and organic dyes due to their chemical stability, good dispersibility in water, low photobleaching and low cytotoxicity. They show great potential for bioimaging, photocatalysis, energy conversion, fluorescent ink and sensing applications [1-3]. In a bioimaging application perspective, the detection of bacteria by microscopic visualization is an essential benchmark. Currently, visual detection approaches are based on indirect methods related to bacterially secreted metabolites or imaging of bacterial colonies [4]. Furthermore, the staining techniques use either commercially available fluorescent dyes or semiconductor quantum dots [5]. Fluorescent dyes are expensive, instable and easily susceptible to photobleaching, while the semiconductor quantum dots are toxic and difficult to dissolve in water. Therefore, simple and inexpensive methods to visualize the morphological details of bacterial cells are highly needed.
C-dots can thus be proposed as an innovative platform for bioimaging purposes thanks to their fluorescent features. In this context, the quantum yield (QY) is one of the most important features for C-dot performance. Although, at present, the actual mechanism of the photoluminescence of C-dots is still an open debate among researchers [6-8], significant progress in increasing the QY has been achieved. Most of the mentioned methods refer to surface passivation [9-11] and doping [12,13]. Recently, chemical reduction was also reported as an effective method to enhance the QY of C-dots [14]. Zheng et al. found an increase in QY for C-dots from 2% to 24% following reduction with sodium borohydride (NaBH4). The same results were confirmed by Shen and Tian's group [15,16]. It was also reported that the fluorescence intensity of graphene quantum dots reduced by hydrazine hydrate (N2H4) can be enhanced to more than two times that of the pristine graphene quantum dots [17]. However, this reduction pathway is based on a two-step procedure: firstly, a synthesis and collection of bare C-dots, then a reduction of C-dots to enhance their QY. The above procedure is often time consuming, poses difficulty in achieving a final pure sample, and introduces secondary pollution products. Therefore, in order to promote and extend their range of applications, new methods to obtain C-dots with high QY are required.
Citric acid, citrate, urea or thiourea have been used in the past to obtain high-QY C-dots with different growth mechanisms proposed [18-20]. Qu et al. obtained graphene quantum dots with a quantum yield of 78% and 71% using citric acid and urea or citric acid and thiourea as the precursors, respectively. They demonstrated that N or N/S doping led to the high QY of the C-dots. Zeng et al. prepared C-dots with a relatively high QY value (45%) using citric acid and urea as precursors via a facile hydrothermal method. They evidenced that surface passivation by urea resulted in the high QY of the C-dots. Herein we report a C-dot synthetic procedure with remarkable QY (37%) by a one-step hydrothermal chemical reduction method, where sodium citrate is the carbon source and urea and thiourea are the co-reducers. Specifically, the amount of citrate was kept constant and the molar ratio of urea and thiourea were varied to demonstrate the effects of thiourea and urea on the different QYs. The results showed C-dots prepared with both urea and thiourea present more reduced carbon and exhibit a higher QY under the synergistic reduction way. Compared with a conventional two-step chemical reduction pathway, the one-step method is efficient and eco-friendly. Moreover, the obtained C-dots with abundant functional groups on the C-dot surface and high QY exhibit excellent potential for use as bacteria (Xanthomonas axonopodis pv. glycins, Xag) imaging agents.
Results and Discussion
Synthesis of carbon nanodots
As shown in Table 1, the C-dots with different QYs were obtained as the amount of sodium citrate was kept constant and the molar ratio of urea and thiourea was varied. Remarkably, C-dots from sodium citrate, urea and thiourea resulted in a higher QY than those of citrate and urea or citrate and thiourea. To explain the differences in QY for these samples, we propose that sodium citrate serves as a self-assembly trigger for a carbon-based structure due to the intermolecular H-bonding. Subsequently, a condensation process takes place, forming C-dots. Meanwhile, the gradual, homogenous release of OH− and NH3 from urea hydrolysis [21] and H2S from thiourea led to the formation of C-dots under alkaline, reducing and hydrothermal conditions [22-25]. Therefore, C-dots prepared with both urea and thiourea present a higher amount of reduced carbon and exhibit a higher QY under the co-reduction pathway. It was also found that increasing the thiourea concentration above 0.014 M during the synthesis process resulted in a gradual decrease in QY. The results indicated that it acquired a highest reduced atmosphere when the molar ratio of urea to thiourea is about 3.
Table 1: Fluorescent carbon nanodots with different quantum yields synthesized using different additives.
| Sample labela | Sodium citrate (mmol) | Urea (mmol) | Thiourea (mmol) | QY (%) |
|---|---|---|---|---|
| Sa | 0.28 | 1.68 | 0 | 8 |
| Sb | 0.28 | 1.26 | 0.42 | 37 |
| Sc | 0.28 | 0.84 | 0.84 | 20 |
| Sd | 0.28 | 0.42 | 1.26 | 14 |
| Se | 0.28 | 0 | 1.68 | 2 |
aAll the samples were dissolved in 30 mL of water.
To demonstrate the rationale of co-reduced C-dot production method, glucose and xylose were used as the model carbon source references. Glucose and xylose were selected due to their broad range of use for C-dot fabrication [26,27] and the obtained QYs are shown in Supporting Information File 1,Table S1. The results further prove that mixed reducing conditions can enhance the QY, irrespective of the carbon source used.
Due to the wide range of C-dot applications, their quantitative yield for mass production must be improved. As shown in Supporting Information File 1,Table S2, 4.0 to 5.0 g of C-dots were prepared by increasing the concentration of the reagents, while still keeping the molar ratios constant during the synthesis. It was found that the QY of the C-dots with co-reduction using urea and thiourea is much higher than when urea or thiourea are used individually during reduction. Moreover, during the scale-up synthesis, it was determined that the C-dots tend to aggregate due to their larger magnitude in mass, leading to an increase in the size of the synthesized C-dots (Figure S1, Supporting Information File 1) and a decrease in the QY.
Characterization of the carbon nanodots
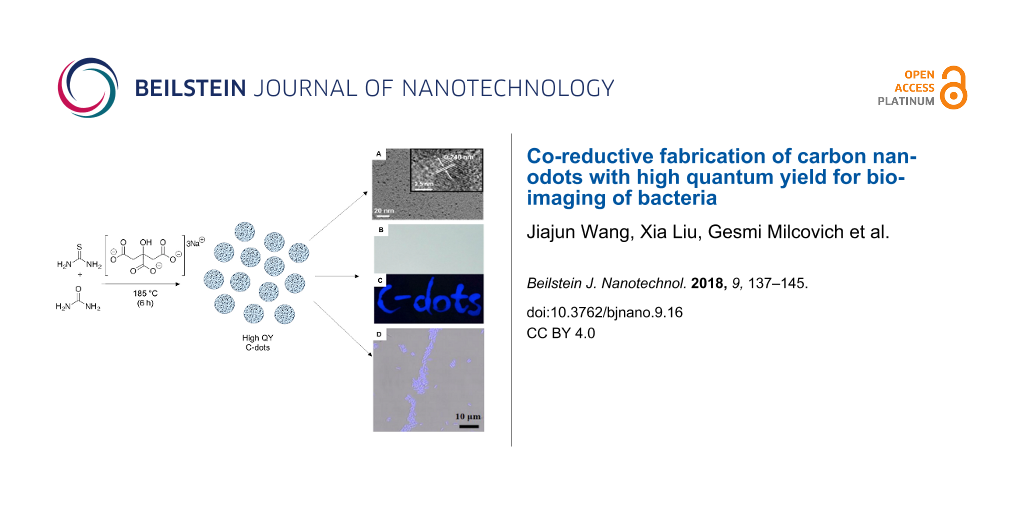
The morphology of the products was characterized by transmission electron microscopy (TEM) and high-resolution transmission electron microscopy (HRTEM). Figure 1A–C shows the TEM images of the Sa, Sb and Se samples. As it can be observed, all the C-dots are consistently dispersed and separated from each other. The three samples show lattice fringes with distances of 0.258 (inset in Figure 1A), 0.246 (inset in Figure 1B) and 0.303 nm (inset in Figure 1C), respectively. These are consistent with the (102), (100) and (002) diffraction planes, respectively, of sp2 graphitic carbon [28,29]. The corresponding particle size distribution histograms (Figure 1D–F) show the average diameter of the Sa, Sb and Se materials is 4.7 ± 1.0 nm, 2.2 ± 0.5 nm and 7.8 ± 1.8 nm, respectively.
![[2190-4286-9-16-1]](/bjnano/content/figures/2190-4286-9-16-1.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 1: TEM and HRTEM (inset) images of (A) Sa, (B) Sb, (C) Se samples, and corresponding size (diameter) distribution ranges for (D) Sa, (E) Sb, and (F) Se.
Figure 1: TEM and HRTEM (inset) images of (A) Sa, (B) Sb, (C) Se samples, and corresponding size (diameter) d...
X-ray diffraction (XRD) was used to investigate the crystallinity of Sa, Sb and Se. As shown in Figure S2 in Supporting Information File 1, sample Sa and Se display a broad diffraction peak centered at around 22.7°, which is similar to the (002) lattice spacing for graphitic (sp2) carbon [30-33]. However, for the pattern of Sb, the peak at 22.7° is much stronger and an extra peak centered at 15.9° appears, which refers to the (103) planes of hexagonal carbon [9]. It can be noticed that the crystallite size from the diffraction peaks does not perfectly match with the lattice spacing observed in TEM. This could be ascribed to the fact that the average polycrystalline signal is collected in XRD, while in TEM, only the single crystallite is investigated.
The mean crystallite size of the C-dots was estimated by using Scherrer’s equation, D = kλ/βcosθ, where D is the average crystallite size, k is a geometrical factor (0.89), λ is the wavelength of the monochromatic X-rays (Cu Kα radiation, λ = 1.5404 Å), θ is the Bragg angle and β is the full-width at half-maximum intensity of the diffraction peak (in radians) at 2θ [34]. The average crystallite size of Sa, Sb and Se calculated from the XRD patterns is 3.3, 2.85 and 3.9 nm, respectively, which is consistent with the size distribution ranges observed in TEM.
In order to further confirm the intrinsic carbogenic structure, the Raman spectra (λex = 633 nm) of Sa, Sb and Se are shown in Figure S3, Supporting Information File 1. Two typical peaks for carbon can be clearly detected for Sa, Sb and Se. The D-band, located at 1387 cm−1, correlates to the disorder or defects in the graphitized structure (sp3-hybridized carbon), while the G-band (1540 cm−1) is assigned to the E2g mode of graphite and corresponds to the vibration of sp2-bonded carbon atoms in a two-dimensional hexagonal [35]. The intensity ratio of the D- to G-band (ID/IG) is a measure of the extent of disorder, and the ratio of sp3/sp2 carbon. Sa has an ID/IG ratio of 0.86, and a ratio of 1 in both Sb and Se was found. The lower ID/IG value of Sb suggests that Sb is composed of more sp2-bonded carbon atoms [36], which agrees well with the proposed co-reduction pathway to effectively produce more reduced carbon with a higher C-dot QY.
UV–vis absorption and fluorescence properties were studied at room temperature to explore the optical properties of the three optimized C-dots. As shown in Figure 2A–C, UV–vis spectra of Sa, Sb and Se all show typical n→π* transition absorption peaks at 336 nm, 332 nm and 330 nm, respectively. Sa, Sb and Se display their highest emission intensity at 430 nm, 420 nm and 430 nm, corresponding to 320 nm, 310 nm and 320 nm excitation, respectively. The emission wavelengths of Sa, Sb and Se are dependent on the excitation wavelengths in the range of 300–380 nm, red-shifting with the gradual increase of the excitation wavelength (Figure 2D–F). Furthermore, the fluorescent decay dynamics for Sa, Sb and Se were also investigated (Figure 2G–I). The time-resolved decay curves of Sa and Sb are well fitted with a tri-exponential function, while that of Se is fitted with a bi-exponential function. The fluorescence characteristic parameters, including the excitation (λex) and emission (λem) wavelengths, quantum yield (Φ), lifetime components (τ1, τ2, τ3) and fluorescence lifetime (τave) for Sa, Sb and Se are listed in Table 2.
![[2190-4286-9-16-2]](/bjnano/content/figures/2190-4286-9-16-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: (A–C) UV–vis absorption, excitation and emission spectra, (D–F) fluorescence emission spectra recorded at different excitation wavelengths and (G–I) fluorescence intensity decay curves of Sa, Sb and Se, respectively.
Figure 2: (A–C) UV–vis absorption, excitation and emission spectra, (D–F) fluorescence emission spectra recor...
Table 2: The excitation (λex) and emission (λem) wavelengths, quantum yield (Φ), lifetime components (τ1, τ2, τ3) and fluorescence lifetime (τave) for Sa, Sb and Se.
| λex (nm) | λem (nm) | Φ | τ1 (ns) | τ2 (ns) | τ3 (ns) | τave (ns) | |
|---|---|---|---|---|---|---|---|
| Sa | 320 | 430 | 8 | 0.08 (1.0%) | 3.98 (19.4%) | 7.61 (79.25%) | 6.17 |
| Sb | 310 | 420 | 37 | 1.06 (2.82%) | 6.19 (73.68) | 10.76 (23.5%) | 7.12 |
| Se | 320 | 430 | 2 | 1.46 (29.3%) | 7.86 (70.7%) | – | 5.98 |
The FT-IR spectra show the functional groups present in the C-dots (Figure 3). All the samples were found to contain oxygen-based functional groups (O–H, C–O, C=O), as well as N–H, C=C, and C–N groups. For Sb and Se, they have additional –NH3+ and sulphur-containing groups, such as S–H and C–S. The weak peak at 1665 cm−1 of Sb indicates an effective reduction of C=O groups.
![[2190-4286-9-16-3]](/bjnano/content/figures/2190-4286-9-16-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: Fourier transform infrared spectra of Sa, Sb, and Se.
Figure 3: Fourier transform infrared spectra of Sa, Sb, and Se.
The chemical composition of the C-dots was further characterized by X-ray photoelectron spectroscopy (XPS) (Figure 4). The C 1s spectra can be fitted by three Gaussian peaks (Figure 4B), which correspond to the sp2-hybridized olefinic carbons (C=C), the sp3-hybridized carbons (C–O, C–S, and C–N), and the oxidized carbon in the carboxyl group. For the Sb sample, the intensity of carboxyl group decreases, whereas the sp2 C=C peak increases. Moreover, a higher binding energy of the graphitic carbon in Sb (284.7 eV) is found compared that of Sa (284.2 eV) and Se (284.3 eV). Increased olefinic sp2 C-bond groups, with shorter bond lengths due to charge neutralization, lead to a stronger interaction between C atoms and higher binding energy. This results in a highly reduced sp2 structure of Sb [37]. The corresponding analytical outcomes are summarized in Table S3, Supporting Information File 1. Thus, Sb exhibits the highest sp2 carbon structure ratio, compared to Sa and Se. According to Figure 4C (N 1s spectra fitting), Sb has the highest intensity of pyridinic-N, suggesting that N atoms are more likely to form pyridinic-N structure during the co-reduction process. The detailed sample data information is presented in Supporting Information File 1, Table S4. The S 2p spectra of Se and Sb reveal the presence of C–S–C (Figure 4D) [38,39]. Overall, XPS and FT-IR results further confirm higher conjugated sp2 C structures in sample Sb, leading to an enhanced fluorescence intensity.
![[2190-4286-9-16-4]](/bjnano/content/figures/2190-4286-9-16-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: XPS spectra of Sa, Sb, and Se. (A) full scan, (B) high-resolution C 1s XPS spectra, (C) high-resolution N 1s XPS spectra, and (D) high-resolution S 2p XPS spectra.
Figure 4: XPS spectra of Sa, Sb, and Se. (A) full scan, (B) high-resolution C 1s XPS spectra, (C) high-resolu...
Carbon nanodots as fluorescent probes for bacteria bioimaging
To explore the potential applications of the high QY C-dots, the highest QY C-dots, Sb, were selected and first utilized to assess their fluorescent characteristics. As shown in Figure 5A (on commercially available filter paper), the characters cannot be detected in the visible wavelength range. Conversely, under UV excitation (λex = 365 nm), the blue fluorescent characters “C-dots” are observed (Figure 5B).
![[2190-4286-9-16-5]](/bjnano/content/figures/2190-4286-9-16-5.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 5: Symbols written on commercially available filter paper using Sb (5.0 μg mL−1) captured under (A) daylight and (B) UV irradiation of λex =365 nm.
Figure 5: Symbols written on commercially available filter paper using Sb (5.0 μg mL−1) captured under (A) da...
After confirming the bright feature behavior, Sb was further used to evaluate its bioimaging properties and bacteria viability range. First of all, a cytotoxicity quantification related to the applicable C-dot concentration range was assessed. Xag viability was evaluated following incubation with Sb in the concentration range from 2.5 to 20 µg mL−1 for 72 h. A positive control (untreated cells) was provided, whereas different time points were designated within the 72 h interval. The data are presented in Figure 6 as the mean ± standard deviation (SD). The results show that no significant cytotoxicity is reported when the concentration of C-dots is lower than 20 µg mL−1. Therefore, C-dots at 20 µg mL−1 concentration are considered as the negative control.
![[2190-4286-9-16-6]](/bjnano/content/figures/2190-4286-9-16-6.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 6: Cytotoxicity towards the bacteria Xag after incubation with Sb in the concentration range 2.5–20 µg mL−1 for 72 h.
Figure 6: Cytotoxicity towards the bacteria Xag after incubation with Sb in the concentration range 2.5–20 µg...
Related to the C-dot assay for bioimaging applications, Xag bacteria were incubated and treated for confocal analysis, as detailed in the Experimental section. According to cytotoxicity assay results, Xag bacteria were incubated with 5.0 μg mL−1 of Sb for 3 h at 37 °C. As shown in Figure 7C, a strong blue fluorescence is observed with λex = 405 nm excitation, whereas no fluorescence signal is detected from the control sample without Sb (Figure 7B). Moreover, both treated and untreated cells appear healthy and consistently preserved, as previously observed in cytotoxicity experiments.
![[2190-4286-9-16-7]](/bjnano/content/figures/2190-4286-9-16-7.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 7: Confocal images of Xag. (A) Bright field without Sb, (B) fluorescence mode without Sb and (C) merged channel image with 5.0 μg mL−1 of Sb incubated for 3 h at 37 °C.
Figure 7: Confocal images of Xag. (A) Bright field without Sb, (B) fluorescence mode without Sb and (C) merge...
Conclusion
High QY (37%) C-dots were synthesized using a direct, simple, one-step reduction reaction process with thiourea and urea as the reducers. Remarkably, the C-dots obtained by the co-reduction showed a highly reduced sp2 structure, bearing a notably high QY. Compared with a conventional two-step chemical reduction pathway, the one-step method is efficient and eco-friendly. Moreover, the obtained C-dots, abundant with the hydrophilic surface, exhibited excellent fluorescent features. On the other hand, a low cytotoxicity for a model bacterial strain Xag was found. Confocal analysis confirmed the suitability of the C-dots as a bioimaging tool for a model bacterial strain, with specific optimal sample synthesis and concentration targeted at this purpose. These unique characteristics look quite promising for the employment of the described systems as fluorescent probes for bioimaging of bacteria Xag.
Experimental
Chemicals and materials
Sodium citrate, urea, and thiourea were purchased from Chemical Reagent Co. Ltd. (Tianjin, China). Xylose and glucose were purchased from Aladdin Ltd. (Shanghai, China). Nutrient broth (NB) medium (1 g L−1 yeast extract, 3 g L−1 beef extract, 5 g L−1 poly peptone, 10 g L−1 sucrose) was purchased from Sigma-Aldrich. All other chemicals were of analytical grade and used as received. Double distilled water was used in all experiments.
Synthesis of carbon nanodots
The C-dots were synthesized by mixing different amounts of thiourea, urea or sodium citrate in 30 mL of distilled water (see Table 1 in Results and Discussion). The aqueous solutions were subsequently transferred to a 50 mL teflon-lined autoclave and heated at a constant temperature of 185 °C for 6 h. After the end of the reaction, suspensions with different turbidity were obtained, evidencing the formation of the C-dots. Then, they were dialyzed against deionized water through a dialyzer with a cut off of Mw = 1000 Da for 40 h. Finally, the resulting solutions were vacuum-evaporated in a rotary evaporator at 50 °C and then freeze-dried to obtain the the C-dot powder. The C-dots were dispersed in ultrapure water as stock solutions (0.5 mg mL−1) for further characterization and use. Table 1 (Results and Discussion) shows the samples with different additives obtained and labeled as Sa, Sb, Sc, Sd, and Se, respectively.
Carbon nanodot characterization
The product morphology was assessed by TEM and HRTEM, which was performed on a JEOL-2100F instrument with an accelerating voltage of 200 kV. The XRD patterns of Sa, Sb, and Se were recorded on a Bruker D8 Advance device with a graphite monochromatized Cu Kα radiation source (λ = 1.54056 Å). XRD diagrams were recorded from 10° to 60° with a step size of 0.02° at 3° min−1. Raman measurements were performed with a Renishaw RM1000 confocal microscope and a He–Ne laser (633 nm, 10 mW). The laser beam was focused to a spot approximately 2 μm in diameter with a 50× microscope objective; the accumulation time was 10 s. Raman spectra were collected from several randomly selected positions on the substrate. Further evidence of the product composition was inferred by means of X-ray photoelectron spectroscopy (XPS), using a K-Alpha XPS spectrometer (Scientific Escalab 250) with an Al Kα X-ray radiation (1486.6 eV) source for excitation. UV–vis absorption spectra of the samples were recorded on a Perkin Elmer Lambda 950 spectrophotometer. FT-IR was conducted at room temperature on a Nicolet 670 spectrometer in the range of 4000–400 cm−1. An LS-45 fluorescence spectrophotometer (Perkin-Elmer, UK) was employed for fluorescence spectroscopy measurements. Confocal images were acquired using a confocal laser scanning microscope (Leica TCS SP5 AOBS).
Fluorescence related experiments
The relative quantum yield was measured according to the equation [40]:
where Φ is the quantum yield, A is the optical density, I is the measured integrated emission intensity, and η is the refractive index. The subscript "std" indicates the value of a standard reference and "x" for the sample. Quinine sulfate (Φstd = 0.54) was used as the standard and was dissolved in 0.1 M H2SO4 (ηstd = 1.33) and the C-dots were dissolved in water (ηx = 1.33). In order to minimize re-absorption effects, absorbance readings in a 10 mm fluorescence cuvette were kept near 0.05 at the excitation wavelength (λex = 360 nm).
Fluorescence lifetime parameters were monitored at room temperature in aqueous solution by using the time-correlated single-photon counting system in the FLS980 device. An Edinburgh EPL 340 ps pulsed diode laser (341.6 nm, 701.2 ps pulse width) operated at 200 kHz was used as the excitation source. The fluorescence lifetime (τ) was fitted by using the Edinburgh FLS980 software package. The average lifetime () was calculated according to the following equation
where Bi is the fractional contribution of the time-resolved decay lifetime of τi.
To further prove and demonstrate the fluorescent features of the C-dots, the colorless aqueous solution of the C-dots (Sb, 5.0 μg mL−1) was applied to commercial filter paper using a Chinese brush. The abbreviation “C-dots” was observed under UV lamp excitation (λex = 365 nm).
Confocal microscopic imaging of bacteria using carbon nanodots as bioimaging probes
Xanthomonas axonopodis pv. glycins (Xag) strains were used as the bacterial model. Xag were grown in NB medium at 28 °C for 24 h. 200 μL of Xag grown in NB medium was then inoculated into 20 mL of fresh medium and grown in a shaking incubator (200 rpm) at 28 °C for 18 h. Then, 5 mL of the bacteria in the middle of an exponential growth phase were collected by centrifugation at 3000 rpm for 10 min (20 °C) and fixed in 1 mL 70% ethanol for 5 min at 4 °C. The fixed bacteria were suspended in NB medium containing C-dots (5.0 μg mL−1) while shaking for 3 h at 28 °C. The final bacteria pellets were washed with PBS, resuspended in 200 μL of PBS, and then further transferred to a glass slide for confocal imaging using an excitation wavelength of 405 nm. All images were acquired at 630× magnification.
Cytotoxicity assay of carbon nanodots for Xag
The cytotoxicity of C-dots toward Xag was measured in NB medium at 28 °C. Different concentrations of C-dots (0, 2.5, 5.0, 10 and 20 μg mL−1) were added into Erlenmeyer flasks and shaken for 2 min at 180 rpm. Furthermore, 0.2 mL of culture broth was collected at different time points (0–72 h) and their optical density was measured at 600 nm in order to calculate the cell viability. Quantification is reported as relative values to the negative control, where the negative control (untreated) is set to 100% viability.
Supporting Information
| Supporting Information File 1:
Additional experimental data.
The general co-reduction method with urea and thiourea to obtain C-dots, the scale-up synthesis of C-dots, TEM and size distribution of Sb50, XRD patterns of Sa, Sb and Se, Raman spectra of Sa, Sb and Se, deconvoluted C 1s XPS spectra of different C-dots with peak area (A) ratios of the sp3 C or oxidized C to the sp2 C and deconvoluted N 1s XPS spectra of different C-dots with A ratios of the pyrrolic N to pyridinic N. |
||
| Format: PDF | Size: 206.8 KB | Download |
References
-
Miao, P.; Han, K.; Tang, Y.; Wang, B.; Lin, T.; Cheng, W. Nanoscale 2015, 7, 1586–1595. doi:10.1039/c4nr05712k
Return to citation in text: [1] -
Zuo, P.; Lu, X.; Sun, Z.; Guo, Y.; He, H. Microchim. Acta 2016, 183, 519–542. doi:10.1007/s00604-015-1705-3
Return to citation in text: [1] -
Yuan, F.; Li, S.; Fan, Z.; Meng, X.; Fan, L.; Yang, S. Nano Today 2016, 11, 565–586. doi:10.1016/j.nantod.2016.08.006
Return to citation in text: [1] -
Padmavathy, T.; Astha, M.; Logan, M. B.; Swadeshmukul, S. Adv. Drug Delivery Rev. 2010, 62, 424–437. doi:10.1016/j.addr.2009.11.014
Return to citation in text: [1] -
Wiliam, W. Y.; Emmanuel, C.; Rebekah, D.; Vicki, L. C. Biochem. Biophys. Res. Commun. 2006, 348, 781–786. doi:10.1016/j.bbrc.2006.07.160
Return to citation in text: [1] -
Zhu, S.; Song, Y.; Zhao, X.; Shao, J.; Zhang, J.; Yang, B. Nano Res. 2015, 8, 355–381. doi:10.1007/s12274-014-0644-3
Return to citation in text: [1] -
Strauss, V.; Margraf, J. T.; Dirian, K.; Syriannis, Z.; Prato, M.; Wessendort, C.; Hirsch, A.; Clark, T.; Guldi, D. M. Angew. Chem., Int. Ed. 2015, 54, 8297–8302. doi:10.1002/anie.201502482
Return to citation in text: [1] -
Strauss, V.; Margraf, J. T.; Dolle, C.; Butz, B.; Nacken, T. J.; Walter, J.; Bauer, W.; Peukert, W.; Spiecker, E.; Clark, T.; Guldi, D. M. J. Am. Chem. Soc. 2014, 136, 17308–17316. doi:10.1021/ja510183c
Return to citation in text: [1] -
Sun, Y.-P.; Zhou, B.; Lin, Y.; Wang, W.; Fernando, K. A. S.; Pathak, P.; Meziani, M. J.; Harruff, B. A.; Wang, X.; Wang, H.; Luo, P. G.; Yang, H.; Kose, M. E.; Chen, B.; Veca, L. M.; Xie, S.-Y. J. Am. Chem. Soc. 2006, 128, 7756–7757. doi:10.1021/ja062677d
Return to citation in text: [1] [2] -
Shen, J.; Zhu, Y.; Chen, C.; Yang, X.; Li, C. Chem. Commun. 2011, 47, 2580–2582. doi:10.1039/c0cc04812g
Return to citation in text: [1] -
Liu, D.-D.; Su, H.; Cao, Q.; Le, X.-Y.; Mao, Z.-W. RSC Adv. 2015, 5, 40588–40594. doi:10.1039/c5ra04544d
Return to citation in text: [1] -
Arcudi, F.; Đorđević, L.; Prato, M. Angew. Chem., Int. Ed. 2016, 55, 2107–2112. doi:10.1002/anie.201510158
Return to citation in text: [1] -
Xu, Q.; Pu, P.; Zhao, J.; Dong, C.; Gao, C.; Chen, Y.; Chen, J.; Liu, Y.; Zhou, H. J. Mater. Chem. A 2015, 3, 542–546. doi:10.1039/c4ta05483k
Return to citation in text: [1] -
Zheng, H.; Wang, Q.; Long, Y.; Zhang, H.; Huang, X.; Zhu, R. Chem. Commun. 2011, 47, 10650–10652. doi:10.1039/C1CC14741B
Return to citation in text: [1] -
Tian, R.; Hu, S.; Wu, L.; Chang, Q.; Yang, J.; Liu, J. Appl. Surf. Sci. 2014, 301, 156–160. doi:10.1016/j.apsusc.2014.02.028
Return to citation in text: [1] -
Shen, R.; Song, K.; Liu, H.; Li, Y.; Liu, H. ChemPhysChem 2012, 13, 3549–3555. doi:10.1002/cphc.201200018
Return to citation in text: [1] -
Feng, Y.; Zhao, J.; Yan, X.; Tang, F.; Xue, Q. Carbon 2014, 66, 334–339. doi:10.1016/j.carbon.2013.09.008
Return to citation in text: [1] -
Bourlinos, A. B.; Stassinopoulos, A.; Anglos, D.; Zboril, R.; Georgakilas, V.; Giannelis, E. P. Chem. Mater. 2008, 20, 4539–4541. doi:10.1021/cm800506r
Return to citation in text: [1] -
Qu, D.; Zheng, M.; Du, P.; Zhou, Y.; Zhang, L.; Li, D.; Tan, H.; Zhao, Z.; Xie, Z.; Sun, Z. Nanoscale 2013, 5, 12272–12277. doi:10.1039/c3nr04402e
Return to citation in text: [1] -
Li, X.; Zhang, S.; Kulinich, S. A.; Liu, Y.; Zeng, H. Sci. Rep. 2014, 4, 4976–4984. doi:10.1038/srep04976
Return to citation in text: [1] -
Sun, Z.; Kim, J. H.; Zhao, Y.; Bijarbooneh, F.; Malgras, V.; Lee, Y.; Kang, Y.-M.; Dou, S. X. J. Am. Chem. Soc. 2011, 133, 19314–19317. doi:10.1021/ja208468d
Return to citation in text: [1] -
Chen, W.; Yan, L.; Bangal, P. R. J. Phys. Chem. C 2010, 114, 19885–19890. doi:10.1021/jp107131v
Return to citation in text: [1] -
Xiong, S.; Zeng, H. C. Angew. Chem., Int. Ed. 2012, 51, 949–952. doi:10.1002/anie.201106826
Return to citation in text: [1] -
Liang, J.; Jiao, Y.; Jaroniec, M.; Qiao, S. Z. Angew. Chem., Int. Ed. 2012, 51, 11496–11500. doi:10.1002/anie.201206720
Return to citation in text: [1] -
Sun, L.; Wang, L.; Tian, C.; Tan, T.; Xie, Y.; Shi, K.; Li, M. RSC Adv. 2012, 2, 4498–4506. doi:10.1039/C2RA01367C
Return to citation in text: [1] -
Zhu, H.; Wang, X.; Li, Y.; Wang, Z.; Yang, F.; Yang, X. Chem. Commun. 2009, 5118–5120. doi:10.1039/b907612c
Return to citation in text: [1] -
Lai, T.; Zheng, E.; Chen, L.; Wang, X.; Kong, L.; You, C.; Ruan, Y.; Weng, X. Nanoscale 2013, 5, 8015–8021. doi:10.1039/c3nr02014b
Return to citation in text: [1] -
Wang, Q.; Zhang, S.; Zhong, Y.; Yang, X.-F.; Li, Z.; Li, H. Anal. Chem. 2017, 89, 1734–1741. doi:10.1021/acs.analchem.6b03983
Return to citation in text: [1] -
Cheng, C.; Shi, Y.; Li, M.; Xing, M.; Wu, Q. Mater. Sci. Eng., C 2017, 79, 473–480. doi:10.1016/j.msec.2017.05.094
Return to citation in text: [1] -
Xiong, Y.; Schneider, J.; Reckmeier, C. J.; Huang, H.; Kasák, P.; Rogach, A. L. Nanoscale 2017, 9, 11730–11738. doi:10.1039/c7nr03648e
Return to citation in text: [1] -
Zhu, S.; Zhao, X.; Song, Y.; Lu, S.; Yang, B. Nano Today 2016, 11, 128–132. doi:10.1016/j.nantod.2015.09.002
Return to citation in text: [1] -
Shi, L.; Yang, J. H.; Zeng, H. B.; Chen, Y. M.; Yang, S. C.; Wu, C.; Zeng, H.; Yoshihito, O.; Zhang, Q. Nanoscale 2016, 8, 14374–14378. doi:10.1039/c6nr00451b
Return to citation in text: [1] -
Kasprzyk, W.; Bednarz, S.; Żmudzki, P.; Galica, M.; Bogdal, D. RSC Adv. 2015, 5, 34795–34799. doi:10.1039/c5ra03226a
Return to citation in text: [1] -
Mohagheghpour, E.; Moztarzadeh, F.; Rabiee, M.; Tahriri, M.; Ashuri, M.; Sameie, H.; Salimi, R.; Moghadas, S. IEEE Trans. NanoBioscience 2012, 11, 317–323. doi:10.1109/TNB.2012.2210442
Return to citation in text: [1] -
Knight, D. S.; White, W. B. J. Mater. Res. 1989, 4, 385–393. doi:10.1557/jmr.1989.0385
Return to citation in text: [1] -
Zhou, X.; Guo, S.; Zhong, P.; Xie, Y.; Li, Z.; Ma, X. RSC Adv. 2016, 6, 54644–54648. doi:10.1039/c6ra06012a
Return to citation in text: [1] -
Gao, F.; Zeng, D.; Huang, Q.; Tian, S.; Xie, C. Phys. Chem. Chem. Phys. 2012, 14, 10572–10578. doi:10.1039/c2cp41045a
Return to citation in text: [1] -
Choi, C. H.; Chung, M. W.; Park, S. H.; Woo, S. I. Phys. Chem. Chem. Phys. 2013, 15, 1802–1805. doi:10.1039/c2cp44147k
Return to citation in text: [1] -
Wohlgemuth, S.-A.; White, R. J.; Willinger, M.-G.; Titirici, M.-M.; Antonietti, M. Green Chem. 2012, 14, 1515–1523. doi:10.1039/c2gc35309a
Return to citation in text: [1] -
Crosby, G. A.; Demas, J. N. J. Phys. Chem. 1971, 75, 991–1024. doi:10.1021/j100678a001
Return to citation in text: [1]
| 36. | Zhou, X.; Guo, S.; Zhong, P.; Xie, Y.; Li, Z.; Ma, X. RSC Adv. 2016, 6, 54644–54648. doi:10.1039/c6ra06012a |
| 34. | Mohagheghpour, E.; Moztarzadeh, F.; Rabiee, M.; Tahriri, M.; Ashuri, M.; Sameie, H.; Salimi, R.; Moghadas, S. IEEE Trans. NanoBioscience 2012, 11, 317–323. doi:10.1109/TNB.2012.2210442 |
| 35. | Knight, D. S.; White, W. B. J. Mater. Res. 1989, 4, 385–393. doi:10.1557/jmr.1989.0385 |
| 1. | Miao, P.; Han, K.; Tang, Y.; Wang, B.; Lin, T.; Cheng, W. Nanoscale 2015, 7, 1586–1595. doi:10.1039/c4nr05712k |
| 2. | Zuo, P.; Lu, X.; Sun, Z.; Guo, Y.; He, H. Microchim. Acta 2016, 183, 519–542. doi:10.1007/s00604-015-1705-3 |
| 3. | Yuan, F.; Li, S.; Fan, Z.; Meng, X.; Fan, L.; Yang, S. Nano Today 2016, 11, 565–586. doi:10.1016/j.nantod.2016.08.006 |
| 9. | Sun, Y.-P.; Zhou, B.; Lin, Y.; Wang, W.; Fernando, K. A. S.; Pathak, P.; Meziani, M. J.; Harruff, B. A.; Wang, X.; Wang, H.; Luo, P. G.; Yang, H.; Kose, M. E.; Chen, B.; Veca, L. M.; Xie, S.-Y. J. Am. Chem. Soc. 2006, 128, 7756–7757. doi:10.1021/ja062677d |
| 10. | Shen, J.; Zhu, Y.; Chen, C.; Yang, X.; Li, C. Chem. Commun. 2011, 47, 2580–2582. doi:10.1039/c0cc04812g |
| 11. | Liu, D.-D.; Su, H.; Cao, Q.; Le, X.-Y.; Mao, Z.-W. RSC Adv. 2015, 5, 40588–40594. doi:10.1039/c5ra04544d |
| 30. | Xiong, Y.; Schneider, J.; Reckmeier, C. J.; Huang, H.; Kasák, P.; Rogach, A. L. Nanoscale 2017, 9, 11730–11738. doi:10.1039/c7nr03648e |
| 31. | Zhu, S.; Zhao, X.; Song, Y.; Lu, S.; Yang, B. Nano Today 2016, 11, 128–132. doi:10.1016/j.nantod.2015.09.002 |
| 32. | Shi, L.; Yang, J. H.; Zeng, H. B.; Chen, Y. M.; Yang, S. C.; Wu, C.; Zeng, H.; Yoshihito, O.; Zhang, Q. Nanoscale 2016, 8, 14374–14378. doi:10.1039/c6nr00451b |
| 33. | Kasprzyk, W.; Bednarz, S.; Żmudzki, P.; Galica, M.; Bogdal, D. RSC Adv. 2015, 5, 34795–34799. doi:10.1039/c5ra03226a |
| 6. | Zhu, S.; Song, Y.; Zhao, X.; Shao, J.; Zhang, J.; Yang, B. Nano Res. 2015, 8, 355–381. doi:10.1007/s12274-014-0644-3 |
| 7. | Strauss, V.; Margraf, J. T.; Dirian, K.; Syriannis, Z.; Prato, M.; Wessendort, C.; Hirsch, A.; Clark, T.; Guldi, D. M. Angew. Chem., Int. Ed. 2015, 54, 8297–8302. doi:10.1002/anie.201502482 |
| 8. | Strauss, V.; Margraf, J. T.; Dolle, C.; Butz, B.; Nacken, T. J.; Walter, J.; Bauer, W.; Peukert, W.; Spiecker, E.; Clark, T.; Guldi, D. M. J. Am. Chem. Soc. 2014, 136, 17308–17316. doi:10.1021/ja510183c |
| 9. | Sun, Y.-P.; Zhou, B.; Lin, Y.; Wang, W.; Fernando, K. A. S.; Pathak, P.; Meziani, M. J.; Harruff, B. A.; Wang, X.; Wang, H.; Luo, P. G.; Yang, H.; Kose, M. E.; Chen, B.; Veca, L. M.; Xie, S.-Y. J. Am. Chem. Soc. 2006, 128, 7756–7757. doi:10.1021/ja062677d |
| 5. | Wiliam, W. Y.; Emmanuel, C.; Rebekah, D.; Vicki, L. C. Biochem. Biophys. Res. Commun. 2006, 348, 781–786. doi:10.1016/j.bbrc.2006.07.160 |
| 26. | Zhu, H.; Wang, X.; Li, Y.; Wang, Z.; Yang, F.; Yang, X. Chem. Commun. 2009, 5118–5120. doi:10.1039/b907612c |
| 27. | Lai, T.; Zheng, E.; Chen, L.; Wang, X.; Kong, L.; You, C.; Ruan, Y.; Weng, X. Nanoscale 2013, 5, 8015–8021. doi:10.1039/c3nr02014b |
| 4. | Padmavathy, T.; Astha, M.; Logan, M. B.; Swadeshmukul, S. Adv. Drug Delivery Rev. 2010, 62, 424–437. doi:10.1016/j.addr.2009.11.014 |
| 28. | Wang, Q.; Zhang, S.; Zhong, Y.; Yang, X.-F.; Li, Z.; Li, H. Anal. Chem. 2017, 89, 1734–1741. doi:10.1021/acs.analchem.6b03983 |
| 29. | Cheng, C.; Shi, Y.; Li, M.; Xing, M.; Wu, Q. Mater. Sci. Eng., C 2017, 79, 473–480. doi:10.1016/j.msec.2017.05.094 |
| 17. | Feng, Y.; Zhao, J.; Yan, X.; Tang, F.; Xue, Q. Carbon 2014, 66, 334–339. doi:10.1016/j.carbon.2013.09.008 |
| 21. | Sun, Z.; Kim, J. H.; Zhao, Y.; Bijarbooneh, F.; Malgras, V.; Lee, Y.; Kang, Y.-M.; Dou, S. X. J. Am. Chem. Soc. 2011, 133, 19314–19317. doi:10.1021/ja208468d |
| 40. | Crosby, G. A.; Demas, J. N. J. Phys. Chem. 1971, 75, 991–1024. doi:10.1021/j100678a001 |
| 15. | Tian, R.; Hu, S.; Wu, L.; Chang, Q.; Yang, J.; Liu, J. Appl. Surf. Sci. 2014, 301, 156–160. doi:10.1016/j.apsusc.2014.02.028 |
| 16. | Shen, R.; Song, K.; Liu, H.; Li, Y.; Liu, H. ChemPhysChem 2012, 13, 3549–3555. doi:10.1002/cphc.201200018 |
| 22. | Chen, W.; Yan, L.; Bangal, P. R. J. Phys. Chem. C 2010, 114, 19885–19890. doi:10.1021/jp107131v |
| 23. | Xiong, S.; Zeng, H. C. Angew. Chem., Int. Ed. 2012, 51, 949–952. doi:10.1002/anie.201106826 |
| 24. | Liang, J.; Jiao, Y.; Jaroniec, M.; Qiao, S. Z. Angew. Chem., Int. Ed. 2012, 51, 11496–11500. doi:10.1002/anie.201206720 |
| 25. | Sun, L.; Wang, L.; Tian, C.; Tan, T.; Xie, Y.; Shi, K.; Li, M. RSC Adv. 2012, 2, 4498–4506. doi:10.1039/C2RA01367C |
| 14. | Zheng, H.; Wang, Q.; Long, Y.; Zhang, H.; Huang, X.; Zhu, R. Chem. Commun. 2011, 47, 10650–10652. doi:10.1039/C1CC14741B |
| 37. | Gao, F.; Zeng, D.; Huang, Q.; Tian, S.; Xie, C. Phys. Chem. Chem. Phys. 2012, 14, 10572–10578. doi:10.1039/c2cp41045a |
| 12. | Arcudi, F.; Đorđević, L.; Prato, M. Angew. Chem., Int. Ed. 2016, 55, 2107–2112. doi:10.1002/anie.201510158 |
| 13. | Xu, Q.; Pu, P.; Zhao, J.; Dong, C.; Gao, C.; Chen, Y.; Chen, J.; Liu, Y.; Zhou, H. J. Mater. Chem. A 2015, 3, 542–546. doi:10.1039/c4ta05483k |
| 18. | Bourlinos, A. B.; Stassinopoulos, A.; Anglos, D.; Zboril, R.; Georgakilas, V.; Giannelis, E. P. Chem. Mater. 2008, 20, 4539–4541. doi:10.1021/cm800506r |
| 19. | Qu, D.; Zheng, M.; Du, P.; Zhou, Y.; Zhang, L.; Li, D.; Tan, H.; Zhao, Z.; Xie, Z.; Sun, Z. Nanoscale 2013, 5, 12272–12277. doi:10.1039/c3nr04402e |
| 20. | Li, X.; Zhang, S.; Kulinich, S. A.; Liu, Y.; Zeng, H. Sci. Rep. 2014, 4, 4976–4984. doi:10.1038/srep04976 |
| 38. | Choi, C. H.; Chung, M. W.; Park, S. H.; Woo, S. I. Phys. Chem. Chem. Phys. 2013, 15, 1802–1805. doi:10.1039/c2cp44147k |
| 39. | Wohlgemuth, S.-A.; White, R. J.; Willinger, M.-G.; Titirici, M.-M.; Antonietti, M. Green Chem. 2012, 14, 1515–1523. doi:10.1039/c2gc35309a |
© 2018 Wang et al.; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Nanotechnology terms and conditions: (http://www.beilstein-journals.org/bjnano)