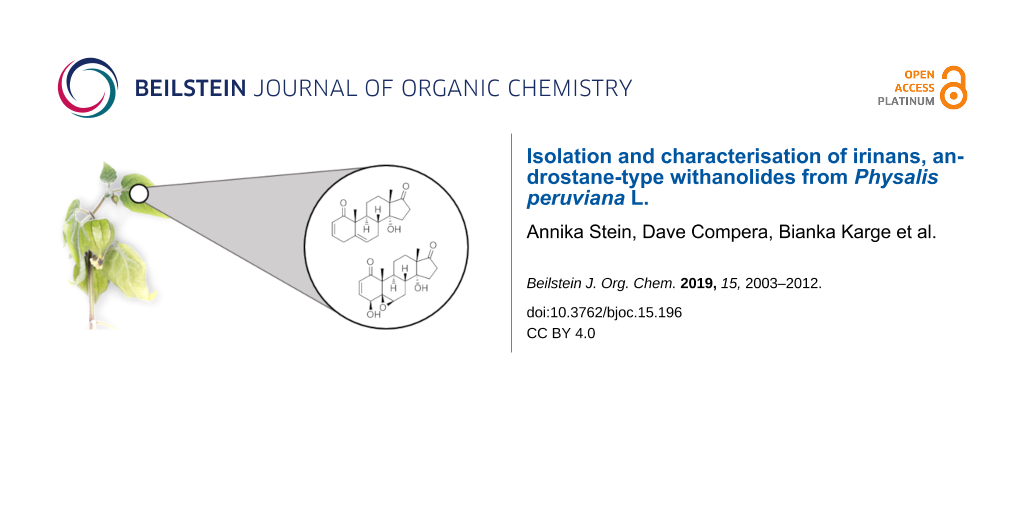
Abstract
Withanolides are steroidal lactones widespread in Nightshade plants with often potent antiproliferative activities. Additionally, the structural diversity of this compound class holds much potential for the discovery of novel biological activity. Here, we report two newly characterised withanolides, named irinans, from Physalis peruviana with highly unusual truncated backbones that resemble mammalian androstane sex hormones. Based on biomimetic chemical reactions, we propose a model that links these compounds to withanolide biosynthesis. Irinans have potent antiproliferative activities, that are however lower than those of 4ß-hydroxywithanolide E. Our work establishes androwithanolides as a new subclass of withanolides.

Graphical Abstract
Introduction
Traditional medicine has long been a source of inspiration for modern drug research. An important example is Withania somnifera, also known as ashwaghanda or Indian ginseng, which has been used in Ayurvedic medicine to treat a large variety of ailments [1]. Extensive studies revealed withanolides, a class of steroidal lactones, to be primarily responsible for the medicinal effects [1,2]. A large range of pharmacological properties has been assigned to withanolides, with antiproliferative activities being the most potent ones [1]. Withanolides have been also discovered in numerous genera other than Withania, for example Datura, Dunalis, Iochroma, Jaborosa, Lycium and Physalis [3], resulting in more than 300 known representatives [3]. Physalis peruviana is a withanolide producer of particular relevance as it is widely cultivated for its edible berries [4]. So far, several withanolides have been reported from P. peruviana and other Physalis species, most prominently physalins, perulactones and 4ß-hydroxywithanolide E (1) [5-16]. As part of our ongoing programme focussed on the biochemistry of withanolides, our aim was to gain further insights into the withanolide profile of P. peruviana. Here we report irinans A (2) and B (3), two unusual truncated withanolides with androstane backbones. We show that oxidative, but not acidic or basic conditions enable conversion of the putative precursor 4ß-hydroxywithanolide E (1) to irinan A (2). Based on this intrinsic reactivity we propose a biosynthetic model that will serve as further guidance for elucidating the enzymatic basis of androstane formation in plants in the future.
Results and Discussion
To isolate withanolides from P. peruviana, we used a purification strategy based on previous reports [17-19]. Nine weeks old whole P. peruviana plants (140 g) were extracted with H2O/MeOH (3:1) and divided into fractions soluble in petroleum ether, chloroform, and n-butanol, respectively. The chloroform fraction was further separated by flash chromatography on a C18 stationary phase, resulting in three major subfractions F1–F3. Final purification by preparative HPLC followed by NMR analysis revealed 4β-hydroxywithanolide E (1) as the major compound (50 mg) as well as the known metabolites withanolide E (4), withanolide F (5) and perulactone H (6) by comparison to literature data (Figure 1) [9,20].
Figure 1: Withanolides from Physalis peruviana. A) Structures of the newly characterised truncated withanolides irinan A (2) and B (3) with an androstane backbone, together with the known withanolides 1 and 4–6 isolated from P. peruviana. B) P. peruviana plant.
Figure 1: Withanolides from Physalis peruviana. A) Structures of the newly characterised truncated withanolid...
Two additional compounds attracted our attention based on their unusual 1H NMR spectra (Table 1). Both showed two multiplets in the olefinic region, which are highly characteristic for withanolides with A-ring Michael acceptors. However, compared to other withanolides, several signals were missing. Typically, withanolides show five singlets for methyl groups in the aliphatic region, as well as the H-22 oxymethine proton of the lactone moiety. Surprisingly, both compounds showed only two putative methyl signals, and no signal which might correspond to H-22. Thus, we reasoned that both unknown compounds might be truncated withanolide-like compounds.
Table 1: 13C and 1H NMR data (CDCl3, 500 MHz, 298 K) of irinans A (2) and B (3) in comparison to the known compound 4β-hydroxywithanolide E (1, CDCl3, 400 MHz, 298 K, δ in ppm, J in Hz). For carbon numbering see Figure 1 and Figure 2.
| 13C | 1H | |||||
|---|---|---|---|---|---|---|
| position | 1 | 2 | 3 | 1 | 2 | 3 |
| 1 | 201.9 | 202.0 | 203.8 | – | – | – |
| 2 | 133.2 | 132.3 | 128.1 | 6.22 (1H, d, 9.9) | 6.22 (1H, d, 10.0) | 5.91 (1H, ddd, 10.0, 3.1, 1.2) |
| 3 | 141.4 | 142.1 | 145.3 | 6.92 (1H, dd, 9.9, 6.1) | 6.94 (1H, dd, 10.0, 5.8) | 6.79 (1H, ddd, 10.0, 5.0, 2.6) |
| 4 | 70.4 | 69.9 | 33.6 | 3.74 (1H, d, 6.1) | 3.79 (1H, dd, 5.8, 2.4) |
3.31 (1H, dddddd, 21.3, 2.8, 2.8, 2.8, 2.8, 2.8)a
2.88 (1H, dd, 21.2, 4.9) |
| 5 | 64.2 | 63.9 | 135.7 | – | – | – |
| 6 | 63.1 | 63.1 | 124.3 | 3.28 (1H, br s) | 3.37 (1H, m) | 5.64 (1H, dt, 5.7, 2.0) |
| 7 | 26.0 | 24.9 | 24.1 | 2.03 (2H, m) |
2.11 (1H, dt, 14.2, 3.1)
1.84 (1H, ddd, 14.1, 11.7, 1.4) |
2.08 (1H, m)
1.95 (1H, m) |
| 8 | 34.3 | 32.6 | 35.5 | 1.83 (1H, m) | 1.90 (1H, m) | 1.88 (1H, m) |
| 9 | 36.7 | 38.1 | 37.1 | 1.69 (1H, m) | 1.51 (1H, m) | 2.10 (1H, m) |
| 10 | 47.9 | 47.8 | 50.9 | – | – | – |
| 11 | 21.5 | 20.4 | 21.6 |
1.72 (1H, m)
1.56 (1H, m) |
1.91 (1H, m)
1.46 (1H, m) |
2.34 (1H, m)
1.52 (1H, m) |
| 12 | 29.8 | 24.3 | 25.0 |
2.25 (1H, m)
1.28 (1H, m) |
1.66 (1H, d, 13.2)
1.55 (1H, m) |
1.86 (1H, m)
1.63 (1H, m) |
| 13 | 54.6 | 52.6 | 52.5 | – | – | – |
| 14 | 81.9 | 80.9 | 81.0 | – | – | – |
| 15 | 32.5 | 30.0 | 29.9 |
1.66 (1H, m)
1.59 (1H, m) |
1.92 (2H, m) | 1.96–1.85 (2H, m) |
| 16 | 38.0 | 33.1 | 33.1 |
2.72 (1H, m)
1.45 (1H, m) |
2.44 (1H, ddd, 18.9, 7.6, 4.1)
2.33 (1H, dt, 18.8, 8.8) |
2.35–2.46 (2H, m) |
| 17 | 87.8 | 218.0 | 218.5 | – | – | – |
| 18 | 20.4 | 17.9 | 18.1 | 1.07 (3H, s) | 1.01 (3H, s) | 1.05 (3H, s) |
| 19 | 16.9 | 17.8 | 19.2 | 1.42 (3H, s) | 1.45 (3H, s) | 1.27 (3H, s) |
| 20 | 79.2 | – | – | – | – | – |
| 21 | 19.8 | – | – | 1.42 (3H, s) | – | – |
| 22 | 79.7 | – | – | 4.88 (1H, dd, 11.8, 5.3) | – | – |
| 23 | 34.4 | – | – | 2.51 (2H, m) | – | – |
| 24 | 150.8 | – | – | – | – | – |
| 25 | 121.6 | – | – | – | – | – |
| 26 | 166.0 | – | – | – | – | – |
| 27 | 12.5 | – | – | 1.88 (3H, s) | – | – |
| 28 | 20.8 | – | – | 1.94 (3H, s) | – | – |
| 14-OH | n.d. | 1.41 (1H, br s) | 1.41 (1H, br s) | |||
| 4-OH | n.d. | 2.57 (1H, d, 2.50) | - | |||
aApparent dsext. See Figure S19 (Supporting Information File 1) for details. n.d. not detected.
HRESIMS suggested a sum formula of C19H24O5 for the first compound, which was supported by the 13C spectrum (Table 1). By comparing the spectrum to NMR data of other withanolides, we quickly identified the Michael system in ring A based on two olefinic protons (δH 6.94 and 6.22 ppm), a secondary alcohol at C-4 (δH 3.79 ppm), a 5,6-epoxide (δH 3.37 (H-6)), and a tertiary alcohol at C-14 (δC 80.9 ppm). COSY correlations supported by HMBC analysis (Figure 2A) revealed an intact ABCD ring system with a substitution pattern identical to 4β-hydroxywithanolide E (1). Only a single, striking difference was noted: C-17 was shifted from 87.8 to 218.0 ppm, strongly suggesting the presence of a ketone instead of an alcohol. In agreement with the predicted sum formula and the absence of all side chain carbons, this completed the structure of the first unknown compound, which we named irinan A (2, Figure 1).
Figure 2: Key NMR correlations. (A) COSY and HMBC correlations for irinan A (2). (B) COSY and HMBC correlations for irinan B (3). (C) Key NOESY correlations for irinan A (2). (D) Key NOESY correlations for irinan B (3).
Figure 2: Key NMR correlations. (A) COSY and HMBC correlations for irinan A (2). (B) COSY and HMBC correlatio...
The second unknown compound had a sum formula of C19H24O3 based on HRESIMS and 13C NMR (Table 1). In contrast to the first compound, no epoxide and no secondary alcohol at C-4 was present, in agreement with the different elemental composition. Instead, 13C NMR indicated a double bond at C5–C6 (δC 135.7 and 124.3 ppm). Otherwise, all spin systems and correlations indicated a typical withanolide ABCD ring system. Again, a carbon with a distinct downfield shift of 218.4 ppm was found, demonstrating the presence of a ketone at C-17. Highly unusually, H-4β appeared as a doublet of sextets (1:5:10:10:5:1) (dsext) in the 1H NMR spectrum. This multiplet was explained as “dddddd” by a total of six COSY correlations (Figure S19, Supporting Information File 1). The resulting compound was named irinan B (3, Figure 1). A compound of putatively identical structure was isolated from P. peruviana before, but only fragmentary physicochemical data has been reported so far [14].
To elucidate the relative stereochemistry of irinans A (2) and B (3), we analysed NOESY data (Figure 2). In the case of irinan A (2), the β configuration of OH-4 was deduced by the NOESY correlation OH-4/CH3-19. OH-14 was assigned as α based on the correlations OH-14/H-12α and H-9/H-12α. The 5,6-epoxide was determined as β by a correlation from H-6 to H-3. These assignments are in complete agreement with the relative stereochemistry of 4ß-hydroxywithanolide E (1). In irinan B, the configuration of OH-14 could not be unambiguously inferred from NOE data due to the signal overlap of H-15 with H-12 and other protons. As an alternative, OH-14 α configuration was deduced from the chemical shifts of C-12 and C-9, which experience a strong shielding γ-gauche effect for OH-14α configurations [21]. These data indicate a relative stereochemistry of irinan B matching withanolide F (5).
Irinans represent highly unusual withanolide derivatives, as they lack the side-chain lactone ring that is a common structural feature of virtually all known withanolides [3], but possess an androstane backbone instead. While androstanes such as androsterone (7) are well-known human sex hormones (Figure 3A) [22], their occurrence in plants is rare [23-26]. Only a single withanolide androstane has been fully characterised before, cinedione (8), isolated from Physalis cinerascens (Figure 3A) [23]. We propose the name androwithanolides for this withanolide subclass, which so far appears to be characteristic of Physalis species.
Figure 3: Structures and biosynthesis of androstanes. (A) Androstane backbone and androsterone (7) as a typical mammalian sex hormone. Cinedione (8) is the only other fully characterised androwithanolide known. (B) Biosynthesis of androstanes in mammals. (C) Possible cleavage mechanisms involved in androwithanolide biosynthesis in plants.
Figure 3: Structures and biosynthesis of androstanes. (A) Androstane backbone and androsterone (7) as a typic...
The biosynthesis of androstanes in mammals requires three enzymatic steps starting from cholesterol (9, Figure 3B) [27]. Cholesterol (9) is converted to pregnenolone (10) by the cytochrome P450 cholesterol side-chain cleavage enzyme (P450scc), which cleaves the C20–C22 bond [27]. Then, the bifunctional P450c17 acts as a 17α-hydroxylase and 17,20-lyase to give rise to androstanes [27]. Related enzymes have not been reported from plants. We searched transcriptome data of P. peruviana for putative homologues of these enzymes [28]. The best hits only had amino acid sequence identities of 22–28%, indicating that no P450 enzymes of these clans exist in P. peruviana. Although enzymes with similar catalytic activity might have evolved convergently in plants, the different substitution pattern in the side chain suggests that a side-chain cleavage mechanism distinct from mammals is involved. While the order of oxidative steps in withanolide biosynthesis is still completely elusive [29], we propose that this fragmentation occurs at a late stage, when most typical withanolide functionalisations have already been introduced. Indeed, irinan A (2), irinan B (3) and cinedione (8) can be directly linked to the known withanolides 4ß-hydroxywithanolide E (1), withanolide F (5) and withanolide S [23], respectively (Figure S20, Supporting Information File 1). If the fragmentation occurred early in the biosynthesis, this would imply that several biosynthetic enzymes have to tolerate substrates without the lactone side chain. We therefore propose that the side-chain cleavage enzyme in withanolide biosynthesis acts at a late stage, using common pathway end products such as 4ß-hydroxywithanolide E (1) as its substrates. Two mechanisms are conceivable for this transformation (Figure 3C): A non-oxidative Grob fragmentation could make use of a push–pull mechanism between C-17 and C-22, building on acid–base catalysis. Alternatively, an enzyme could cleave the C17–C20 diol oxidatively. Several P450 enzymes have been reported to be capable of cleaving diols, presumably via a ferric peroxo intermediate (Figure 3C) [30,31].
To gain further insights into the biosynthetic route and to exclude that androwithanolides are isolation artefacts [32], we exposed 4ß-hydroxywithanolide E (1) as the likely precursor to irinan A (2) to various chemical conditions (Figure 4). In general, 1 was stable in all solvents tested, namely chloroform, methanol, DMSO and acetonitrile (data not shown). Treatment with acid at pH 3 caused no reaction at all when heating up to 70 °C (Figure 4A). At pH 0, several unidentified compounds appeared, but not irinan A (2). In basic conditions, only a single unidentified product was formed at pH 11 and 70 °C. Next, we tested whether 1 could be oxidatively cleaved [33]. Incubation of 1 with NaIO4 at room temperature did not result in any reaction (data not shown). However, although it has been reported that periodates are not capable of cleaving ditertiary glycols [33,34], we noted formation of small quantities of irinan A (2) when performing the reaction at 70 °C (Figure 4B and Figure S21 in Supporting Information File 1). The identity of irinan A (2) was verified by isolation of the corresponding compound by preparative HPLC (4% yield) followed by NMR analysis. This result confirms our NMR-based stereochemical assignment and unambiguously links irinan A (2) to 4ß-hydroxywithanolide E (1). We also performed an oxidative cleavage reaction with catalytic amounts of MoO2(acac)2 in DMSO as described by García et al. [34], which also led to the formation of trace amounts of irinan A (2). Our experiments suggest that irinan A (2) and most likely all androwithanolides are not isolation artefacts but true natural products, which require an oxidative enzyme to facilitate the C–C bond cleavage. Future studies will shed light on the enzymatic basis of androwithanolide formation.
![[1860-5397-15-196-4]](/bjoc/content/figures/1860-5397-15-196-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: Intrinsic reactivity of 4ß-hydroxywithanolide E (1) under acidic/basic and oxidative conditions, respectively. (A) LC–MS chromatograms (ELS detection) of 1 incubated at different pH values. (B) LC–MS chromatograms (UV detection at 200–400 nm) of 1 treated with different oxidative reagents. The formation of 2 in the NaIO4 reaction was confirmed by NMR analysis. See also Figure S21 in Supporting Information File 1 for extracted ion chromatograms (EICs).
Figure 4: Intrinsic reactivity of 4ß-hydroxywithanolide E (1) under acidic/basic and oxidative conditions, re...
Considering the potent bioactivities of androstanes as well as withanolides, we wondered whether the loss of the side-chain lactone would negatively impact the antiproliferative activity. Irinan A (2) and B (3) together with 4ß-hydroxywithanolide E (1) as a positive control were evaluated against a panel of four cell lines (Table 2). In our assays we observed decreasing activities during the third and fourth replicates, resulting in large standard deviations and potentially indicating limited stability of these compounds. Nonetheless, EC50 values of 4ß-hydroxywithanolide E (1) were in good agreement with previously published values [14,35,36]. Irinans A (2) and B (3) were 1.3 to 10-fold less active than 4ß-hydroxywithanolide E (1), with the exception of irinan B (3) in A549 cells, which was equipotent. However, irinan A (2) and B (3) samples had a purity of 90% and 80%, respectively. We therefore cannot exclude that unidentified impurities, which could not be removed by repeated preparative HPLC, obscure the true EC50 values of irinans. We conclude that irinans possess potent antiproliferative activity, that is however reduced compared to 4ß-hydroxywithanolide E (1). Our results demonstrate the importance of the lactone side chain for bioactivity.
Table 2: Antiproliferative activities in different cell lines. Data indicate EC50 values ± SD in µM. A549 = human lung carcinoma; L929 = mouse fibroblast; KB-3-1 = human cervix carcinoma; MCF-7 = human breast cancer cell line.
| Compound | A549 | L929 | KB-3-1 | MCF-7 |
|---|---|---|---|---|
| 4ß-hydroxywithanolide E (1) | 3.74 ± 0.50 | 0.27 ± 0.30 | 1.11 ± 0.98 | 10.65 ± 6.18 |
| irinan A (2)a | 5.01 ± 5.27 | 2.29 ± 0.88 | 4.62 ± 5.76 | 17.88 ± 7.27 |
| irinan B (3)b | 3.45 ± 1.91 | 1.68 ± 1.78 | 2.40 ± 2.32 | 13.56 ± 9.18 |
| staurosporine (positive control) | 1.19 ± 0.99 | <0.003 | 0.04 ± 0.01 | 0.16 ± 0.02 |
| auranofin (positive control) | >7.03 | 2.35 ± 0.83 | 1.59 ± 0.37 | 2.06 ± 0.60 |
aEstimated 90% purity based on 1H NMR. bEstimated 80% purity based on 1H NMR.
Conclusion
We have discovered and characterised irinans A and B, two new withanolides from P. peruviana with truncated backbones. They resemble mammalian sex hormones of the androstane class. The relative stereochemistry was elucidated based on NOESY analysis. Chemical studies support a model that these compounds are formed by an oxidative process. We propose the name androwithanolides for this withanolide subclass.
Experimental
General experimental procedures
Seeds of Physalis peruviana were obtained from FloraSelf, Sperli and Quedlinburger Saatgut. Plants were initially grown in seed starter soil (Kölle’s Beste Anzuchterde) and later transferred to potting soil (Kölle’s Beste Pflanzerde). Plants were grown under LED illumination (SANlight S2W) at 350 µmol s−1 m−2 PPFD with a 12 h photoperiod and at 18–25 °C without temperature and humidity control. Plants were watered twice per week with tap water as needed.
NMR spectra were recorded using Bruker AscendTM 400 or DRX 500 MHz spectrometers operating at 400 and 500 MHz for 1H NMR and at 100 and 125 MHz for 13C NMR where CDCl3 was used as solvent. Chemical shifts were referenced relative to the residual solvent signal of CDCl3 (δH = 7.26 ppm, δC = 77.16 ppm) and expressed in δ values (ppm), with coupling constants reported in Hz. Analysis was conducted with TopSpin (Version 4.0.6, Bruker). ATR-IR analysis was performed for the range of 400–4000 cm−1 using a Shimadzu IRAffinity 1S spectrometer with samples dissolved in chloroform. Optical rotations were measured with a Perkin Elmer 341 polarimeter. Using methanol as solvent, the wavelength for maximum absorption was determined on a Jasco V-630 spectrophotometer. Flash purification was performed on a Biotage Isolera One using columns described below. HRMS measurements were carried out on a Waters Alliance 2695 HPLC coupled to a Micromass LCT Premier mass spectrometer.
For analytical and preparative LC–MS a Waters instrument was used consisting of a Waters 2767 autosampler, Waters 2545 pump system, Waters 2998 diode array detector, Waters 2424 ELS detector, and a Waters SQ Detector 2 for mass spectrometry in ESI+ and ESI– modes between m/z 150 and 1000. In analytical mode, a Phenomenex Kinetex column (2.6 µm, C18, 100 Å, 4.6 × 100 mm) was used with a gradient of [solvent A: H2O + 0.05% formic acid; solvent B: acetonitrile + 0.045% formic acid; gradient: 10% to 90% B over 10 min, 1 mL/min]. Samples were dissolved to a concentration of 10 mg/mL in MeOH and 20 µL injected. In preparative mode, a Phenomenex Kinetex Axia column (5 µm, C18, 100 Å, 21.2 × 250 mm) equipped with a Phenomenex Security Guard precolumn (Luna, C5, 300 Å) was used in combination with the separation gradient described below.
Extraction and isolation of withanolides
140 g of 9 weeks old, whole Physalis peruviana plants were frozen in liquid nitrogen and ground to a fine powder. The powder was extracted with 500 mL H2O/MeOH (3:1) at room temperature for 3 h. After filtration and evaporation of the solvent under reduced pressure, the crude extract was resuspended in 300 mL H2O and defatted with 300 mL petroleum ether. The remaining aqueous layer was further extracted with 2 × 300 mL CHCl3 followed by 2 × 300 mL n-BuOH. This resulted in a 660 mg petroleum ether fraction, 386 mg CHCl3 fraction and 1174 mg n-BuOH fraction.
The CHCl3 fraction was separated via reversed-phase flash chromatography (Biotage SNAP KP-C18-HS 30 g column) with a H2O/MeOH gradient. Samples were adsorbed onto Celite under reduced pressure for dry loading. A gradient from 30% to 95% MeOH was used. Fractions were pooled guided by UV maximum absorbance to form main fraction F1 (subfractions 1–21, 102 mg), F2 (subfractions 22–31, 12 mg) and F3 (subfractions 32–45, 48 mg). No withanolides were detected in F2 based on LC–MS analysis and therefore discarded.
Fraction F1 was further separated by preparative LC–MS. The sample was dissolved in MeOH to a concentration of 15 mg/mL. 100 µL was injected per run. A separation gradient was used [solvent A: H2O + 0.05% formic acid; solvent B: acetonitrile + 0.045% formic acid; gradient: 10% to 90% B over 10 min, 20 mL/min]. The post-column flow was split (100:1) and the minority flow made up to 1 mL/min with MeOH + 0.045% formic acid for in-line analysis by UV, ELSD and MS. The majority flow was collected. The following peaks were collected and identified by NMR: tR = 5.8–6.0 min (irinan A (2), 6 mg); 7.2–8.0 min (4β-hydroxywithanolide E (1), 49 mg); 9.0–9.2 min (irinan B (3), 1 mg). The collected fractions were evaporated under reduced pressure using a Christ RVC 2-25 CDplus rotational vacuum concentrator.
Main fraction F3 was also separated by preparative LC–MS as described above, yielding the known compounds withanolide E (4) (tR = 7.3–7.6 min, 6 mg), perulactone H (6) (tR = 7.6–7.8 min, 9 mg) and withanolide F (5) (tR = 7.8–8.5 min, 9 mg) which were identified by NMR [20].
Analytical data
4β-Hydroxywithanolide E (1) was isolated as a white crystalline powder. NMR data of 1 is listed in Table 1. All spectroscopic properties matched literature data [20].
Irinan A (2): white crystalline powder; [α]D20 +10.48 (β = 0.62; MeOH); UV (MeOH) λmax (log ε) 239 nm (3.93); IR (ATR, CHCl3) νmax: 3460, 2967, 2930, 1734, 1674, 1454, 1373, 1092, 1036, 986, 922, 754 cm−1; for 1H and 13C data see Table 1; HRESIMS m/z: [M + Na]+ calcd for C19H24O5Na+, 355.1516; found, 355.1519.
Irinan B (3): white crystalline powder; [α]D20 –10.00 (β = 0.06; MeOH); UV (MeOH) λmax (log ε) 251 nm (3.94); IR (ATR, CHCl3) νmax: 3402, 2955, 2930, 1682, 1383, 1259, 1215, 1136, 1088, 1016, 966, 806, 748 cm−1; for 1H and 13C data see Table 1; HRESIMS m/z: [M + Na]+ calcd for C19H24O3Na+, 323.1618; found, 323.1626
BLAST search of known androstane biosynthesis enzymes
The known androstane biosynthesis enzymes Homo sapiens P450scc (UniProtKB accession P05108) and Homo sapiens P450c17 (P05093) were used to search reported Physalis peruviana transcriptome data [28] via the tBLASTn algorithm. Both enzymes yielded several full-length hits with amino acid sequence identities of 22–28%.
Oxidative cleavage of 4β-hydroxywithanolide E (1) to irinan A (2) by NaIO4
57.8 mg of NaIO4 (270.2 µmol, 7.0 equiv) in 400 µL hot H2O was added to 19.4 mg 4ß-hydroxywithanolide E (1, 38.6 µmol, 1.0 equiv) in 1 mL MeOH. The reaction was incubated at 70 °C for 72 h in a heat block with shaking at 1000 rpm. After that time a peak with m/z 315 corresponding to [M + H − H2O]+ with a retention time of 3.5 min was observed by LC–MS, co-eluting with authentic irinan A (2). The reaction mixture was separated by preparative LC–MS as described above to give a white crystalline powder (0.5 mg, 4%), which was confirmed to be irinan A (2) by 1H NMR spectroscopy.
Oxidative cleavage of 4β-hydroxywithanolide E (1) to irinan A (2) by MoO2(acac)2
2 µL of a MoO2(acac)2 stock solution in DMSO (100 µg/µL, 0.6 µmol, 0.02 equiv) was added to 14.6 mg hydroxywithanolide E (1, 29.0 µmol, 1.0 equiv) in 100 µL DMSO. The reaction was incubated at 130 °C for 3 h in an oil bath with stirring at 400 rpm. After that time a peak with m/z 315 corresponding to [M + H − H2O]+ with a retention time of 3.5 min was observed, co-eluting with authentic irinan A (2). H2O (5 mL) and CHCl3 (5 mL) were added to the reaction mixture. The layers were separated and the aqueous phase was extracted with chloroform (3 × 5 mL). Combined organic layers were washed with water (5 mL), dried over MgSO4, filtered and the solvent was evaporated under reduced pressure. The resulting crude reaction product was then analysed by analytical HPLC as described above.
Antiproliferative assays
The effect of compounds on cell viability was probed with a WST-1 test using the procedure of Ishiyama et al. [37] as modified by Sasse et al. [38]. The following cell lines were used: mouse fibroblast cell line L929 (DSM ACC 2), human cervix carcinoma cell line KB-3-1 (DSM ACC 158), the human lung carcinoma cell line A549 (DSMZ ACC 107) and human breast cancer cell line MCF-7 (DSM ACC 115). The subconfluent cells were briefly washed with Earle’s Balanced Salt Solution (Gibco) without Ca and Mg, trypsinized and re-suspended in Dulbecco’s modified eagle’s medium that contained 5% fetal bovine serum (FBS; L929, KB-3-1, A549) or Roswell Park Memorial Institute medium that contained 5% FBS, 0.5% Minimum Essential Medium Non-Essential Amino Acids, Gibco (MEM NEAA), 0.5% GlutaMAX (Gibco) and insulin at 5 μg/mL (MCF-7). 25 µL of serial dilutions of the test compounds (64–0.06 µg/mL, that were made with a pipetting robot (epMotion, Eppendorf, Hamburg, Germany), were added to 25 μL aliquots of a cell suspension (1500 cells for KB-3-1, L929 and A549, 3000 cells for MCF-7) in 384 well microtiter plates. Blank and solvent controls were incubated under identical conditions. After an incubation period of 5 days, 3 μL WST-1 (ready to use solution by Roche) was added. The incubation time of the plates at 37 °C varied between the cell lines from 20 min for KB-3-1 and A549, L929 for 30 min, and 2 h for MCF-7 before measuring absorbance at 450 nm (reference 600 nm) with an Infinite 200 PRO plate reader (Tecan, Männedorf, Switzerland). As positive control compounds, Auranofin and Staurosporin were applied. The absorbance of the solvent control was set to 100%. The EC50 values were determined with Sigma Plot. All data are average values from four biological replicates.
Supporting Information
| Supporting Information File 1: NMR, MS, UV and IR spectra of irinan A (2) and irinan B (3). NMR data of withanolide E (4), withanolide F (5) and perulactone H (6). | ||
| Format: PDF | Size: 931.9 KB | Download |
Acknowledgements
This work has been carried out within the framework of the SMART BIOTECS alliance between the Technische Universität Braunschweig and the Leibniz Universität Hannover. This initiative is supported by the Ministry of Science and Culture (MWK) of Lower Saxony, Germany. LC–MS (INST 187/621) and NMR (INST 187/686-1) instruments funded by the Deutsche Forschungsgemeinschaft (DFG) were used. We thank Prof. Russell Cox for his support and helpful discussions, and Marcel Arndt for preliminary work. We thank Dr. Jörg Fohrer and colleagues for support with NMR measurements and Katja Körner and colleagues for excellent lab support.
References
-
Rai, M.; Jogee, P. S.; Agarkar, G.; Santos, C. A. d. Pharm. Biol. (Abingdon, U. K.) 2016, 54, 189–197. doi:10.3109/13880209.2015.1027778
Return to citation in text: [1] [2] [3] -
Maurya, R. J. Pharm. Pharmacol. 2010, 62, 153–160. doi:10.1211/jpp.62.02.0001
Return to citation in text: [1] -
Chen, L.-X.; He, H.; Qiu, F. Nat. Prod. Rep. 2011, 28, 705–740. doi:10.1039/c0np00045k
Return to citation in text: [1] [2] [3] -
Fischer, G.; Herrera, A.; Almanza, P. J. Cape Gooseberry (Physalis Peruviana L.). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E. M., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Elsevier: Amsterdam, Netherlands, 2011; pp 374–397. doi:10.1533/9780857092762.374
Return to citation in text: [1] -
Sang-ngern, M.; Youn, U. J.; Park, E.-J.; Kondratyuk, T. P.; Simmons, C. J.; Wall, M. M.; Ruf, M.; Lorch, S. E.; Leong, E.; Pezzuto, J. M.; Chang, L. C. Bioorg. Med. Chem. Lett. 2016, 26, 2755–2759. doi:10.1016/j.bmcl.2016.04.077
Return to citation in text: [1] -
Park, E.-J.; Sang-Ngern, M.; Chang, L. C.; Pezzuto, J. M. J. Nat. Prod. 2019, 82, 492–499. doi:10.1021/acs.jnatprod.8b00861
Return to citation in text: [1] -
Kirson, I.; Abraham, A.; Sethi, P. D.; Subramanian, S. S.; Glotter, E. Phytochemistry 1976, 15, 340–342. doi:10.1016/s0031-9422(00)89029-9
Return to citation in text: [1] -
Frolow, F.; Ray, A. B.; Sahai, M.; Glotter, E.; Gottlieb, H. E.; Kirson, I. J. Chem. Soc., Perkin Trans. 1 1981, 1029–1032. doi:10.1039/p19810001029
Return to citation in text: [1] -
Sakurai, K.; Ishii, H.; Kobayashi, S.; Iwao, T. Chem. Pharm. Bull. 1976, 24, 1403–1405. doi:10.1248/cpb.24.1403
Return to citation in text: [1] [2] -
Neogi, P.; Sahai, M.; Ray, A. B. Phytochemistry 1986, 26, 243–247. doi:10.1016/s0031-9422(00)81520-4
Return to citation in text: [1] -
Dinan, L. N.; Sarker, S. D.; Šik, V. Phytochemistry 1997, 44, 509–512. doi:10.1016/s0031-9422(96)00553-5
Return to citation in text: [1] -
Fang, S.-T.; Liu, J.-K.; Li, B. Steroids 2012, 77, 36–44. doi:10.1016/j.steroids.2011.09.011
Return to citation in text: [1] -
Ahmad, S.; Malik, A.; Yasmin, R.; Ullah, N.; Gul, W.; Khan, P. M.; Nawaz, H. R.; Afza, N. Phytochemistry 1999, 50, 647–651. doi:10.1016/s0031-9422(98)00567-6
Return to citation in text: [1] -
Lan, Y.-H.; Chang, F.-R.; Pan, M.-J.; Wu, C.-C.; Wu, S.-J.; Chen, S.-L.; Wang, S.-S.; Wu, M.-J.; Wu, Y.-C. Food Chem. 2009, 116, 462–469. doi:10.1016/j.foodchem.2009.02.061
Return to citation in text: [1] [2] [3] -
Fang, S.-T.; Li, B.; Liu, J.-K. Helv. Chim. Acta 2009, 92, 1304–1308. doi:10.1002/hlca.200900005
Return to citation in text: [1] -
Zhang, W.-N.; Tong, W.-Y. Chem. Biodiversity 2016, 13, 48–65. doi:10.1002/cbdv.201400435
Return to citation in text: [1] -
Xia, G.; Li, Y.; Sun, J.; Wang, L.; Tang, X.; Lin, B.; Kang, N.; Huang, J.; Chen, L.; Qiu, F. Steroids 2016, 115, 136–146. doi:10.1016/j.steroids.2016.09.002
Return to citation in text: [1] -
Xu, Y.-m.; Bunting, D. P.; Liu, M. X.; Bandaranayake, H. A.; Gunatilaka, A. A. L. J. Nat. Prod. 2016, 79, 821–830. doi:10.1021/acs.jnatprod.5b00911
Return to citation in text: [1] -
Chen, L.-X.; Xia, G.-Y.; He, H.; Huang, J.; Qiu, F.; Zi, X.-L. RSC Adv. 2016, 6, 52925–52936. doi:10.1039/c6ra07031k
Return to citation in text: [1] -
Ozawa, M.; Morita, M.; Hirai, G.; Tamura, S.; Kawai, M.; Tsuchiya, A.; Oonuma, K.; Maruoka, K.; Sodeoka, M. ACS Med. Chem. Lett. 2013, 4, 730–735. doi:10.1021/ml400144e
Return to citation in text: [1] [2] [3] -
Zhang, H.; Timmermann, B. N. J. Nat. Prod. 2016, 79, 732–742. doi:10.1021/acs.jnatprod.5b00648
Return to citation in text: [1] -
Kicman, A. T. Br. J. Pharmacol. 2008, 154, 502–521. doi:10.1038/bjp.2008.165
Return to citation in text: [1] -
Maldonado, E.; Alvarado, V. E.; Torres, F. R.; Martínez, M.; Pérez-Castorena, A. L. Planta Med. 2005, 71, 548–553. doi:10.1055/s-2005-864157
Return to citation in text: [1] [2] [3] -
Siddiqui, B. S.; Usmani, S. B.; Begum, S.; Siddiqui, S. Phytochemistry 1993, 33, 925–928. doi:10.1016/0031-9422(93)85306-c
Return to citation in text: [1] -
Sanogo, R.; Germano, M. P.; de Tommasi, N.; Pizza, C.; Aquino, R. Phytochemistry 1998, 47, 73–78. doi:10.1016/s0031-9422(97)00477-9
Return to citation in text: [1] -
Pupo, M. T.; Vieira, P. C.; Fernandes, J. B.; das G.F. da Silva, M. F.; Fo, E. R. Phytochemistry 1997, 45, 1495–1500. doi:10.1016/s0031-9422(97)00167-2
Return to citation in text: [1] -
Miller, W. L.; Auchus, R. J. Endocr. Rev. 2011, 32, 81–151. doi:10.1210/er.2010-0013
Return to citation in text: [1] [2] [3] -
Fukushima, A.; Nakamura, M.; Suzuki, H.; Yamazaki, M.; Knoch, E.; Mori, T.; Umemoto, N.; Morita, M.; Hirai, G.; Sodeoka, M.; Saito, K. Front. Plant Sci. 2016, 7, 1883. doi:10.3389/fpls.2016.01883
Return to citation in text: [1] [2] -
Dhar, N.; Razdan, S.; Rana, S.; Bhat, W. W.; Vishwakarma, R.; Lattoo, S. K. Front. Plant Sci. 2015, 6, 1031. doi:10.3389/fpls.2015.01031
Return to citation in text: [1] -
Strushkevich, N.; MacKenzie, F.; Cherkesova, T.; Grabovec, I.; Usanov, S.; Park, H.-W. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 10139–10143. doi:10.1073/pnas.1019441108
Return to citation in text: [1] -
Ortiz de Montellano, P. R. Substrate Oxidation by Cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Ortiz de Montellano, P. E., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp 111–176. doi:10.1007/978-3-319-12108-6_4
Return to citation in text: [1] -
Capon, R. J. Nat. Prod. Rep. 2019. doi:10.1039/c9np00013e
Return to citation in text: [1] -
Zviely, M.; Goldman, A.; Kirson, I.; Glotter, E. J. Chem. Soc., Perkin Trans. 1 1986, 229–231. doi:10.1039/p19860000229
Return to citation in text: [1] [2] -
García, N.; Rubio-Presa, R.; García-García, P.; Fernández-Rodríguez, M. A.; Pedrosa, M. R.; Arnáiz, F. J.; Sanz, R. Green Chem. 2016, 18, 2335–2340. doi:10.1039/c5gc02862k
Return to citation in text: [1] [2] -
Yen, C.-Y.; Chiu, C.-C.; Chang, F.-R.; Chen, J. Y.-F.; Hwang, C.-C.; Hseu, Y.-C.; Yang, H.-L.; Lee, A. Y.-L.; Tsai, M.-T.; Guo, Z.-L.; Cheng, Y.-S.; Liu, Y.-C.; Lan, Y.-H.; Chang, Y.-C.; Ko, Y.-C.; Chang, H.-W.; Wu, Y.-C. BMC Cancer 2010, 10, 46. doi:10.1186/1471-2407-10-46
Return to citation in text: [1] -
Xu, Y.-M.; Wijeratne, E. M. K.; Babyak, A. L.; Marks, H. R.; Brooks, A. D.; Tewary, P.; Xuan, L.-J.; Wang, W.-Q.; Sayers, T. J.; Gunatilaka, A. A. L. J. Nat. Prod. 2017, 80, 1981–1991. doi:10.1021/acs.jnatprod.6b01129
Return to citation in text: [1] -
Ishiyama, M.; Tominaga, H.; Shiga, M.; Sasamoto, K.; Ohkura, Y.; Ueno, K. Biol. Pharm. Bull. 1996, 19, 1518–1520. doi:10.1248/bpb.19.1518
Return to citation in text: [1] -
Sasse, F.; Steinmetz, H.; Schupp, T.; Petersen, F.; Memmert, K.; Hofmann, H.; Heusser, C.; Brinkmann, V.; Matt, P. V.; Höfle, G.; Reichenbach, H. J. Antibiot. 2002, 55, 543–551. doi:10.7164/antibiotics.55.543
Return to citation in text: [1]
| 20. | Ozawa, M.; Morita, M.; Hirai, G.; Tamura, S.; Kawai, M.; Tsuchiya, A.; Oonuma, K.; Maruoka, K.; Sodeoka, M. ACS Med. Chem. Lett. 2013, 4, 730–735. doi:10.1021/ml400144e |
| 20. | Ozawa, M.; Morita, M.; Hirai, G.; Tamura, S.; Kawai, M.; Tsuchiya, A.; Oonuma, K.; Maruoka, K.; Sodeoka, M. ACS Med. Chem. Lett. 2013, 4, 730–735. doi:10.1021/ml400144e |
| 28. | Fukushima, A.; Nakamura, M.; Suzuki, H.; Yamazaki, M.; Knoch, E.; Mori, T.; Umemoto, N.; Morita, M.; Hirai, G.; Sodeoka, M.; Saito, K. Front. Plant Sci. 2016, 7, 1883. doi:10.3389/fpls.2016.01883 |
| 1. | Rai, M.; Jogee, P. S.; Agarkar, G.; Santos, C. A. d. Pharm. Biol. (Abingdon, U. K.) 2016, 54, 189–197. doi:10.3109/13880209.2015.1027778 |
| 3. | Chen, L.-X.; He, H.; Qiu, F. Nat. Prod. Rep. 2011, 28, 705–740. doi:10.1039/c0np00045k |
| 23. | Maldonado, E.; Alvarado, V. E.; Torres, F. R.; Martínez, M.; Pérez-Castorena, A. L. Planta Med. 2005, 71, 548–553. doi:10.1055/s-2005-864157 |
| 3. | Chen, L.-X.; He, H.; Qiu, F. Nat. Prod. Rep. 2011, 28, 705–740. doi:10.1039/c0np00045k |
| 27. | Miller, W. L.; Auchus, R. J. Endocr. Rev. 2011, 32, 81–151. doi:10.1210/er.2010-0013 |
| 1. | Rai, M.; Jogee, P. S.; Agarkar, G.; Santos, C. A. d. Pharm. Biol. (Abingdon, U. K.) 2016, 54, 189–197. doi:10.3109/13880209.2015.1027778 |
| 1. | Rai, M.; Jogee, P. S.; Agarkar, G.; Santos, C. A. d. Pharm. Biol. (Abingdon, U. K.) 2016, 54, 189–197. doi:10.3109/13880209.2015.1027778 |
| 2. | Maurya, R. J. Pharm. Pharmacol. 2010, 62, 153–160. doi:10.1211/jpp.62.02.0001 |
| 23. | Maldonado, E.; Alvarado, V. E.; Torres, F. R.; Martínez, M.; Pérez-Castorena, A. L. Planta Med. 2005, 71, 548–553. doi:10.1055/s-2005-864157 |
| 24. | Siddiqui, B. S.; Usmani, S. B.; Begum, S.; Siddiqui, S. Phytochemistry 1993, 33, 925–928. doi:10.1016/0031-9422(93)85306-c |
| 25. | Sanogo, R.; Germano, M. P.; de Tommasi, N.; Pizza, C.; Aquino, R. Phytochemistry 1998, 47, 73–78. doi:10.1016/s0031-9422(97)00477-9 |
| 26. | Pupo, M. T.; Vieira, P. C.; Fernandes, J. B.; das G.F. da Silva, M. F.; Fo, E. R. Phytochemistry 1997, 45, 1495–1500. doi:10.1016/s0031-9422(97)00167-2 |
| 9. | Sakurai, K.; Ishii, H.; Kobayashi, S.; Iwao, T. Chem. Pharm. Bull. 1976, 24, 1403–1405. doi:10.1248/cpb.24.1403 |
| 20. | Ozawa, M.; Morita, M.; Hirai, G.; Tamura, S.; Kawai, M.; Tsuchiya, A.; Oonuma, K.; Maruoka, K.; Sodeoka, M. ACS Med. Chem. Lett. 2013, 4, 730–735. doi:10.1021/ml400144e |
| 21. | Zhang, H.; Timmermann, B. N. J. Nat. Prod. 2016, 79, 732–742. doi:10.1021/acs.jnatprod.5b00648 |
| 17. | Xia, G.; Li, Y.; Sun, J.; Wang, L.; Tang, X.; Lin, B.; Kang, N.; Huang, J.; Chen, L.; Qiu, F. Steroids 2016, 115, 136–146. doi:10.1016/j.steroids.2016.09.002 |
| 18. | Xu, Y.-m.; Bunting, D. P.; Liu, M. X.; Bandaranayake, H. A.; Gunatilaka, A. A. L. J. Nat. Prod. 2016, 79, 821–830. doi:10.1021/acs.jnatprod.5b00911 |
| 19. | Chen, L.-X.; Xia, G.-Y.; He, H.; Huang, J.; Qiu, F.; Zi, X.-L. RSC Adv. 2016, 6, 52925–52936. doi:10.1039/c6ra07031k |
| 3. | Chen, L.-X.; He, H.; Qiu, F. Nat. Prod. Rep. 2011, 28, 705–740. doi:10.1039/c0np00045k |
| 5. | Sang-ngern, M.; Youn, U. J.; Park, E.-J.; Kondratyuk, T. P.; Simmons, C. J.; Wall, M. M.; Ruf, M.; Lorch, S. E.; Leong, E.; Pezzuto, J. M.; Chang, L. C. Bioorg. Med. Chem. Lett. 2016, 26, 2755–2759. doi:10.1016/j.bmcl.2016.04.077 |
| 6. | Park, E.-J.; Sang-Ngern, M.; Chang, L. C.; Pezzuto, J. M. J. Nat. Prod. 2019, 82, 492–499. doi:10.1021/acs.jnatprod.8b00861 |
| 7. | Kirson, I.; Abraham, A.; Sethi, P. D.; Subramanian, S. S.; Glotter, E. Phytochemistry 1976, 15, 340–342. doi:10.1016/s0031-9422(00)89029-9 |
| 8. | Frolow, F.; Ray, A. B.; Sahai, M.; Glotter, E.; Gottlieb, H. E.; Kirson, I. J. Chem. Soc., Perkin Trans. 1 1981, 1029–1032. doi:10.1039/p19810001029 |
| 9. | Sakurai, K.; Ishii, H.; Kobayashi, S.; Iwao, T. Chem. Pharm. Bull. 1976, 24, 1403–1405. doi:10.1248/cpb.24.1403 |
| 10. | Neogi, P.; Sahai, M.; Ray, A. B. Phytochemistry 1986, 26, 243–247. doi:10.1016/s0031-9422(00)81520-4 |
| 11. | Dinan, L. N.; Sarker, S. D.; Šik, V. Phytochemistry 1997, 44, 509–512. doi:10.1016/s0031-9422(96)00553-5 |
| 12. | Fang, S.-T.; Liu, J.-K.; Li, B. Steroids 2012, 77, 36–44. doi:10.1016/j.steroids.2011.09.011 |
| 13. | Ahmad, S.; Malik, A.; Yasmin, R.; Ullah, N.; Gul, W.; Khan, P. M.; Nawaz, H. R.; Afza, N. Phytochemistry 1999, 50, 647–651. doi:10.1016/s0031-9422(98)00567-6 |
| 14. | Lan, Y.-H.; Chang, F.-R.; Pan, M.-J.; Wu, C.-C.; Wu, S.-J.; Chen, S.-L.; Wang, S.-S.; Wu, M.-J.; Wu, Y.-C. Food Chem. 2009, 116, 462–469. doi:10.1016/j.foodchem.2009.02.061 |
| 15. | Fang, S.-T.; Li, B.; Liu, J.-K. Helv. Chim. Acta 2009, 92, 1304–1308. doi:10.1002/hlca.200900005 |
| 16. | Zhang, W.-N.; Tong, W.-Y. Chem. Biodiversity 2016, 13, 48–65. doi:10.1002/cbdv.201400435 |
| 37. | Ishiyama, M.; Tominaga, H.; Shiga, M.; Sasamoto, K.; Ohkura, Y.; Ueno, K. Biol. Pharm. Bull. 1996, 19, 1518–1520. doi:10.1248/bpb.19.1518 |
| 4. | Fischer, G.; Herrera, A.; Almanza, P. J. Cape Gooseberry (Physalis Peruviana L.). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E. M., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Elsevier: Amsterdam, Netherlands, 2011; pp 374–397. doi:10.1533/9780857092762.374 |
| 14. | Lan, Y.-H.; Chang, F.-R.; Pan, M.-J.; Wu, C.-C.; Wu, S.-J.; Chen, S.-L.; Wang, S.-S.; Wu, M.-J.; Wu, Y.-C. Food Chem. 2009, 116, 462–469. doi:10.1016/j.foodchem.2009.02.061 |
| 38. | Sasse, F.; Steinmetz, H.; Schupp, T.; Petersen, F.; Memmert, K.; Hofmann, H.; Heusser, C.; Brinkmann, V.; Matt, P. V.; Höfle, G.; Reichenbach, H. J. Antibiot. 2002, 55, 543–551. doi:10.7164/antibiotics.55.543 |
| 28. | Fukushima, A.; Nakamura, M.; Suzuki, H.; Yamazaki, M.; Knoch, E.; Mori, T.; Umemoto, N.; Morita, M.; Hirai, G.; Sodeoka, M.; Saito, K. Front. Plant Sci. 2016, 7, 1883. doi:10.3389/fpls.2016.01883 |
| 27. | Miller, W. L.; Auchus, R. J. Endocr. Rev. 2011, 32, 81–151. doi:10.1210/er.2010-0013 |
| 27. | Miller, W. L.; Auchus, R. J. Endocr. Rev. 2011, 32, 81–151. doi:10.1210/er.2010-0013 |
| 34. | García, N.; Rubio-Presa, R.; García-García, P.; Fernández-Rodríguez, M. A.; Pedrosa, M. R.; Arnáiz, F. J.; Sanz, R. Green Chem. 2016, 18, 2335–2340. doi:10.1039/c5gc02862k |
| 14. | Lan, Y.-H.; Chang, F.-R.; Pan, M.-J.; Wu, C.-C.; Wu, S.-J.; Chen, S.-L.; Wang, S.-S.; Wu, M.-J.; Wu, Y.-C. Food Chem. 2009, 116, 462–469. doi:10.1016/j.foodchem.2009.02.061 |
| 35. | Yen, C.-Y.; Chiu, C.-C.; Chang, F.-R.; Chen, J. Y.-F.; Hwang, C.-C.; Hseu, Y.-C.; Yang, H.-L.; Lee, A. Y.-L.; Tsai, M.-T.; Guo, Z.-L.; Cheng, Y.-S.; Liu, Y.-C.; Lan, Y.-H.; Chang, Y.-C.; Ko, Y.-C.; Chang, H.-W.; Wu, Y.-C. BMC Cancer 2010, 10, 46. doi:10.1186/1471-2407-10-46 |
| 36. | Xu, Y.-M.; Wijeratne, E. M. K.; Babyak, A. L.; Marks, H. R.; Brooks, A. D.; Tewary, P.; Xuan, L.-J.; Wang, W.-Q.; Sayers, T. J.; Gunatilaka, A. A. L. J. Nat. Prod. 2017, 80, 1981–1991. doi:10.1021/acs.jnatprod.6b01129 |
| 33. | Zviely, M.; Goldman, A.; Kirson, I.; Glotter, E. J. Chem. Soc., Perkin Trans. 1 1986, 229–231. doi:10.1039/p19860000229 |
| 33. | Zviely, M.; Goldman, A.; Kirson, I.; Glotter, E. J. Chem. Soc., Perkin Trans. 1 1986, 229–231. doi:10.1039/p19860000229 |
| 34. | García, N.; Rubio-Presa, R.; García-García, P.; Fernández-Rodríguez, M. A.; Pedrosa, M. R.; Arnáiz, F. J.; Sanz, R. Green Chem. 2016, 18, 2335–2340. doi:10.1039/c5gc02862k |
| 30. | Strushkevich, N.; MacKenzie, F.; Cherkesova, T.; Grabovec, I.; Usanov, S.; Park, H.-W. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 10139–10143. doi:10.1073/pnas.1019441108 |
| 31. | Ortiz de Montellano, P. R. Substrate Oxidation by Cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Ortiz de Montellano, P. E., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp 111–176. doi:10.1007/978-3-319-12108-6_4 |
| 29. | Dhar, N.; Razdan, S.; Rana, S.; Bhat, W. W.; Vishwakarma, R.; Lattoo, S. K. Front. Plant Sci. 2015, 6, 1031. doi:10.3389/fpls.2015.01031 |
| 23. | Maldonado, E.; Alvarado, V. E.; Torres, F. R.; Martínez, M.; Pérez-Castorena, A. L. Planta Med. 2005, 71, 548–553. doi:10.1055/s-2005-864157 |
© 2019 Stein et al.; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0). Please note that the reuse, redistribution and reproduction in particular requires that the authors and source are credited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (https://www.beilstein-journals.org/bjoc)