Abstract
Sesquiterpene synthases in Trichoderma viride have been seldom studied, despite the efficiency of filamentous fungi for terpenoid production. Using the farnesyl diphosphate-overexpressing Saccharomyces cerevisiae platform to produce diverse terpenoids, we herein identified an unknown sesquiterpene synthase from T. viride by genome mining and determined the structure of its corresponding products. One new 5/6 bicyclic sesquiterpene and its esterified derivative were characterised by GC–MS and 1D and 2D NMR spectroscopy. To the best of our knowledge, this is the first well-identified sesquiterpene synthase from T. viride to date.

Graphical Abstract
Introduction
Terpenoids represent the most diverse group of natural products, with a wide distribution in microorganisms, plants, insects and various marine invertebrates [1,2]. More than 80,000 terpenoids have been identified and characterised [3-5]. These diverse and complex natural products are mostly derived from carbocation cyclisation with linear C5 isoprene precursors, which are catalysed by terpene synthases (TPSs) [6]. TPSs can be classified into three types based on their amino acid sequence. Type I TPSs are metal-dependent enzymes that initiate cyclisation by the elimination of diphosphate groups from precursors and carbocation formation, and type II TPSs initiate the catalytic process by the protonation of an olefinic double bond [7]. The recently reported type III TPSs, UbiA-related TPSs, also catalyse cascade reactions by diphosphate elimination [8]. In addition, each type of TPS is characterised by a unique aspartate-rich motif; most type I TPSs have a DDXXD/E motif and an NSE/DTE motif, whereas type II TPSs have the DXDD motif [9,10].
The C15 sesquiterpenoids constitute a large class of terpenoids with a wide range of industrial and commercial applications, including uses in flavours and perfumes, as bioactive molecules in the pharmaceutical industry, and in health care products [11]. Sesquiterpenoids are biosynthesised from the universal linear precursor farnesyl pyrophosphate (FPP) and assembled by FPP synthases, using dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) as substrates. The subsequent elimination of diphosphate from FPP is catalysed by sesquiterpene synthases, with further cyclisation steps to form structurally diverse (poly)cyclic core skeletons [3,12]. A set of post-modification enzymes can transform core sesquiterpene skeletons into different kinds of sesquiterpenoids with potential anticancer, cytotoxic and antibiotic functions [13]. More than 121 skeleton structures derived from the sesquiterpene precursor FPP via sesquiterpene synthase have been described. Nearly 75% of these structures have at least one six-membered ring; 69% of these contain five-membered rings, occupying a large portion. Three- and seven-membered ring structures account for just 21% and 24% of these structures, respectively. Four (10%) and eight (7%) membered ring structures (e.g. asteriscanolide) are seldom found [14]. With the lower costs of gene sequencing, recent developments in genome mining by sequencing and annotation have led to the discovery of a large number of functionally unknown terpene synthases [15-17], generating diverse complex structures and several bioactive products (e.g., 6α,9α,15-trihydroxycadinan-4-en-3-one, (+)-3,11,12-trihydroxycalamenene, and (−)-3,10,11,12-tetrahydroxycalamenene) [18].
Filamentous fungi are powerful producers of terpenoid products [19]. Many terpenoids produced by these fungi have recently been characterised; these terpenoids exhibit diverse complex structures and uncommon catalytic mechanisms [20]. However, a limited number of sesquiterpenes have been characterised from a few fungal taxa (e.g., trefolane A and sterhirsutins) [21]. Trichoderma viride is a filamentous fungus that has received considerable attention as an effective biocontrol agent against two fungal pathogens, Fusarium oxysporum f. sp. adzuki and Pythium arrhenomanes, infecting soybean. This fungus is a competent mycoparasite and strong producer of secondary metabolites [22,23]. However, T. viride terpenoids have rarely been studied and the low concentrations of products under natural conditions have limited the pace of research in this field. Metabolic engineering makes the overproduction of different terpenoids from T. viride possible [21,24]. To increase the discovery efficiency of terpenoid products, heterologous expression of various sources of terpene synthases in Escherichia coli and Saccharomyces cerevisiae is a feasible approach [25,26].
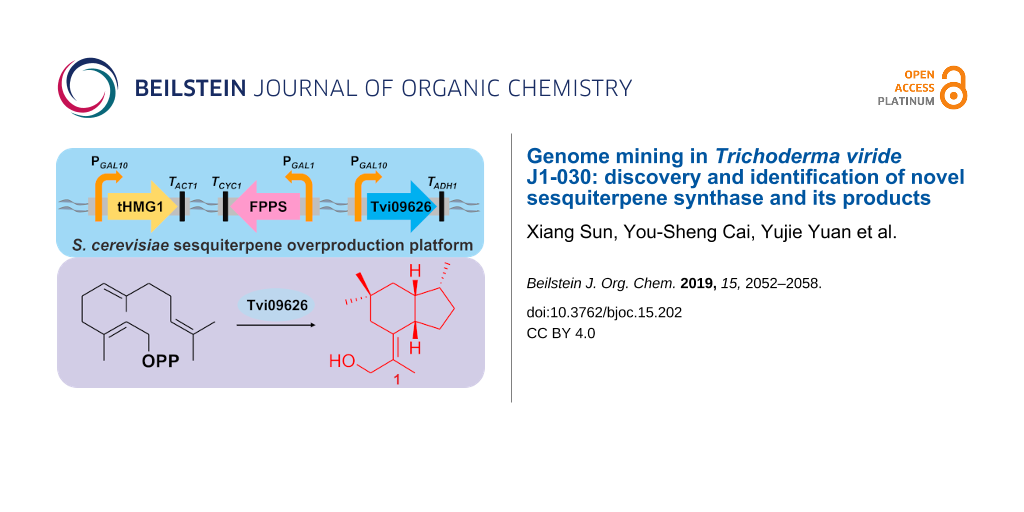
In this study, a combination of genome mining and metabolic engineering was used for sesquiterpenoid discovery, utilizing farnesyl diphosphate-overexpressing S. cerevisiae as a platform (Figure 1). By the heterologous expression of predicted terpene synthases from the genome of T. viride, an unknown sesquiterpene synthase was identified and characterised. Furthermore, a new compound produced by this enzyme and its esterified product were detected and characterised by GC–MS and 1D and 2D NMR, revealing a 5/6 bicyclic sesquiterpene and its C-11 esterified structure. Based on a literature search, to our knowledge, this is the first report of the characterisation of a sesquiterpene synthase in T. viride. In addition, this study demonstrates the effectiveness of the combination of genome mining and heterologous expression of predicted terpene synthases for detecting unknown terpenoids from rarely studied fungi.
![[1860-5397-15-202-1]](/bjoc/content/figures/1860-5397-15-202-1.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 1: Schematic diagram of the S. cerevisiae sesquiterpene overproduction platform and the products of Tvi09626.
Figure 1: Schematic diagram of the S. cerevisiae sesquiterpene overproduction platform and the products of Tv...
Results and Discussion
Prediction and analysis of terpene synthase genes in T. viride J1-030
Through genome sequencing of T. viride J1-030 and prediction of the potential terpene synthases in J1-030 genome, gene Tvi09626 was selected and the following bioinformatics analysis of the function of this unidentified terpene synthase was performed. A protein blast search against the NCBI database was performed with Tvi09626, revealing sequence identities of 89.66% and 85.23% with the enzymes from the strain T. virens Gv29-8 [27] and T. reesei QM6a [28], respectively, with only predicted functions. Thereafter, an amino acid sequence alignment with several known terpene synthases showed that Tvi09626 had the typical highly conserved 128DDxxD/E aspartate-rich motif, 276NSE/DTE triad, 366RY dimer and 230R monomer (Figure S1, Supporting Information File 1) [29-31]. Furthermore, in a phylogenetic analysis (Figure 2), Tvi09626 belonged to Clade V of Class I terpene synthases. In previous studies, terpene synthases have been studied in the genus Trichoderma, such as trichodiene synthase homologous gene isolation and characterisation in T. harzianum [32] and functional identification of terpene synthase vir4 in T. virens [33]. However, owing to the general lack of previous studies of terpene synthases in T. viride, Tvi09626 is the first terpene synthase well-identified with products characterised in T. viride.
![[1860-5397-15-202-2]](/bjoc/content/figures/1860-5397-15-202-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Phylogenetic analysis of Tvi09626 with other characterised terpene synthases. Six clades are marked with different colours and Tvi09626 is labelled in red in Clade V. Percentages indicate branch support based on 1,000 bootstrap replicates.
Figure 2: Phylogenetic analysis of Tvi09626 with other characterised terpene synthases. Six clades are marked...
In vitro analysis of Tvi09626 function
To confirm the function of the candidate enzyme, the DNA sequence of Tvi09626 was amplified by touchdown PCR from the T. viride genome. The gene fragment was cloned into a pET28a (+) vector to construct the plasmid pXS222. Next, pXS222 was transformed into BL21 to overexpress and purify Tvi09626 (Figure S2, Supporting Information File 1). The substrates GPP, FPP and GGPP were incubated with the protein individually and the products were detected and analysed by gas chromatography/mass spectrometry (GC–MS) [30,34]. In vitro assays clearly showed that Tvi09626 could use FPP as its only substrate to produce compound 1 (Figure 3 and Figure S3, Supporting Information File 1).
![[1860-5397-15-202-3]](/bjoc/content/figures/1860-5397-15-202-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: GC–MS chromatogram of products in vivo (I), in yeast YZL141 (II), in vitro Tvi09626 with FPP (III), and boiled Tvi09626 with FPP (IV).
Figure 3: GC–MS chromatogram of products in vivo (I), in yeast YZL141 (II), in vitro Tvi09626 with FPP (III),...
Heterologous expression of Tvi09626 in S. cerevisiae
To further verify the function of the putative terpene synthase, a metabolic engineering strategy was used to reconstruct an FPP overproduction platform in S. cerevisiae in order to obtain sufficient quantities of the products of Tvi09626 for chemical structural characterisation. S. cerevisiae YZL141, engineered previously [21], was used owing to its ability to provide enough IPP, DMAPP, and FPP for the production of terpenoids (Figure 1). After 72 h of shaken-flask fermentation, the strain was extracted with hexane/ethyl acetate (4:1), pre-separated by silica gel column chromatography, and detected by GC–MS. Similar to the in vitro assay results, compound 1 was the final product (Figure 4). Interestingly, during the extraction process, compound 2 was detected at a retention time of 14.53 min and identified as esterified compound 1 (Figure 4 and Table S4, Supporting Information File 1). Using the metabolic engineering strategy, the product of Tvi09626 was efficiently enriched via an abundant FPP supply.
![[1860-5397-15-202-4]](/bjoc/content/figures/1860-5397-15-202-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: Characterisation of Tvi09626 products. (A) Mass spectra of compound 1 at tR = 13.46 min with m/z 222 and compound 2 at tR = 14.53 min with m/z 264. (B) COSY, HMBC and NOESY correlations for compound 1.
Figure 4: Characterisation of Tvi09626 products. (A) Mass spectra of compound 1 at tR = 13.46 min with m/z 22...
Detection and characterisation of Tvi09626 products
By semi-preparative high-performance liquid chromatography (HPLC), we purified compound 1 and compound 2 (23.1 mg and 13.2 mg, respectively). The structures of the two new compounds were characterised by 1D and 2D NMR spectroscopy (Table 1, Table S4, and Figures S4–S15, Supporting Information File 1).
Table 1: 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz) data for compound 1 in CDCl3.
| Position | δC | δH |
|---|---|---|
| 1 | 45.2 | 1.68 (dd, 8.2, 4.3 Hz, 1H) |
| 2 | 40.6 | 1.38 (ddd, J = 12.8, 3.9, 1.6 Hz, 1H), 1.16 (t, J = 12.9 Hz,1H) |
| 3 | 33 | — |
| 4 | 43.8 | 2.35 (dd, J = 13.8, 1.4 Hz, 1H), 1.60 (d, J = 13.8 Hz, 1H) |
| 5 | 137.4 | — |
| 6 | 47.6 | 1.96 (dd, J = 12.3, 6.1 Hz, 1H) |
| 7 | 30.9 | 2.10 (m, 1H), 1.60 (m, 1H) |
| 8 | 33.3 | 2.07 (m, 1H), 1.07 (m, 1H) |
| 9 | 31.6 | 1.99 (m, 1H) |
| 10 | 126.5 | — |
| 11 | 65.2 | 4.18 (d, J = 11.6 Hz, 1H),3.97 (d, J = 9.8 Hz, 1H) |
| 12 | 17.85 | 1.87 (t, J = 1.2 Hz, 3H) |
| 13 | 32 | 0.96 (s, 3H) |
| 14 | 26.1 | 0.83 (s, 3H) |
| 15 | 17.78 | 0.80 (d, J = 7.0 Hz, 3H) |
| 16 | — | 3.47 (s, 1H) |
Compound 1 was a new compound with a known skeleton [35], isolated as a white powder. 1H and 13C NMR data showed four methyl groups at δH 1.87 (t, J = 1.2 Hz, 3H,), δH 0.96 (s, 3H), δH 0.83 (s, 3H), and δH 0.80 (d, J = 7.0 Hz, 3H). Five methylenes were detected, including an oxygenated one at δH 4.18 (d, J = 11.6 Hz, 1H), 3.97 (d, J = 9.8 Hz, 1H), as well as three methines and three quaternary carbons including a double bond at δC 137.4 (C-5), 126.5 (C-10). The 2D NMR data indicated that compound 1 is a 5/6 bicyclic sesquiterpene with the molecular formula C15H26O (Figure 1). Interestingly, compound 1 contained a quaternary carbon with two methyl groups, which is uncommon for the cyclization mechanism of sesquiterpenoids and needs further investigation.
Compound 2 was purified as a white powder. 1H and 13C NMR data showed chemical shifts of five methyl groups at δH 1.83 (t, J = 1.2 Hz, 3H), δH 0.96 (s, 3H), δH 0.82 (s, 3H), δH 0.80 (d, J = 7.0 Hz, 3H), and δH 2.06 (s, 3H). Five methylenes were identified, including an esterified group at C-11 with a resonance of δH 4.64 (d, J = 11.6 Hz 1H), 4.46 (d, J = 11.6 Hz, 1H), three methines, and three quaternary carbons including a double bond at δC 140.37 (C-5), 122.04 (C-10). Compared with 2D NMR information of compound 1, compound 2 was a C-11 esterified 1 with the molecular formula C17H28O2 (Table S4, Supporting Information File 1), which may represent the esterification reaction during the extraction process.
The 5/6 bicyclic sesquiterpene identified and characterised in this study was a brasilane-type sesquiterpenoid; this sesquiterpenoid type is typically isolated from cultures of the basidiomycete Coltricia sideroides in combination with its two new alkane derivatives colisiderin A and (7E,9E)-undeca-7,9-diene-2,4,5-triol [35] and from the organic extract of the red alga Laurencia obtusa [36]. However, to the best of our knowledge, ours is the first report of these brasilane-type sesquiterpenes obtained via biosynthetic genes.
Metal ion dependency of Tvi09626 and its kinetics
As reported previously, most terpene synthases are active in the presence of Mg2+ ions [8,37]. To test the Mg2+ dependency of Tvi09626, an in vitro assay was performed. The GC–MS analysis showed that in the presence of Mg2+, compound 1 can be obtained, whereas without Mg2+ or added EDTA (2.5 mM), compound 1 cannot be detected (Figure 5). This assay demonstrated that Tvi09626 was a Mg2+-dependent sesquiterpene synthase. In a kinetics analysis, the turnover rate (kcat) of the enzyme with FPP was (15 ± 0.3) × 10−2, which is similar to those of omp6 and omp7. Its substrate affinity (Km) was (0.44 ± 0.11) × 10−6, one-tenth that of omp6 and nearly a quarter that of omp7. The catalytic efficiency (kcat/Km) of Tvi09626 was (35.32 ± 0.57) × 103, higher than that of omp6 and lower than that of omp7 [21,38].
![[1860-5397-15-202-5]](/bjoc/content/figures/1860-5397-15-202-5.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 5: GC–MS chromatogram for the metal ion dependency assay.
Figure 5: GC–MS chromatogram for the metal ion dependency assay.
Conclusion
In conclusion, we identified a novel sesquiterpene synthase, Tvi09626, from T. viride using a strong sesquiterpene overproduction platform with S. cerevisiae YZL141; it is the first biochemically identified and characterised sesquiterpene synthase from this filamentous fungus. In an analysis of its relative structure, the product of this enzyme was characterised as a 5/6 bicyclic sesquiterpene compound 1 oxygenated at C-11. Interestingly, esterified compound 1 was isolated during the product extraction process, suggesting an esterification reaction. It contained a quaternary carbon with two methyl groups, which is uncommon of the cyclization mechanism of sesquiterpenoids and needs to be studied in the future. To the best of our knowledge, this study reports the first use of a biosynthetic gene to obtain a brasilane-type sesquiterpene.
Supporting Information
| Supporting Information File 1: Experimental part and supplementary figures and tables. | ||
| Format: PDF | Size: 2.1 MB | Download |
Acknowledgements
We thank Dr. You-Sheng Cai (Wuhan University) for suggestions regarding structural characterisation. This work was financially supported by funding from the National Key R&D Program of China (2018YFA0900400), the Medical Science Advancement Program (Clinical Medicine) of Wuhan University, the state Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (grant MMLKF 18-12) and the National Natural Science Foundation of China (31800032).
References
-
Bian, G. K.; Ma, T.; Liu, T. G. Methods Enzymol. 2018, 608, 97–129. doi:10.1016/bs.mie.2018.04.025
Return to citation in text: [1] -
Huber, T.; Weisheit, L.; Magauer, T. Beilstein J. Org. Chem. 2015, 11, 2521–2539. doi:10.3762/bjoc.11.273
Return to citation in text: [1] -
Christianson, D. W. Chem. Rev. 2017, 117, 11570–11648. doi:10.1021/acs.chemrev.7b00287
Return to citation in text: [1] [2] -
Pemberton, T. A.; Chen, M.; Harris, G. G.; Chou, W. K. W.; Duan, L.; Köksal, M.; Genshaft, A. S.; Cane, D. E.; Christianson, D. W. Biochemistry 2017, 56, 2010–2023. doi:10.1021/acs.biochem.7b00137
Return to citation in text: [1] -
Lauterbach, L.; Rinkel, J.; Dickschat, J. S. Angew. Chem., Int. Ed. 2018, 57, 8280–8283. doi:10.1002/anie.201803800
Return to citation in text: [1] -
Huang, A. C.; Hong, Y. J.; Bond, A. D.; Tantillo, D. J.; Osbourn, A. Angew. Chem., Int. Ed. 2018, 57, 1291–1295. doi:10.1002/anie.201711444
Return to citation in text: [1] -
Bian, G.; Rinkel, J.; Wang, Z.; Lauterbach, L.; Hou, A.; Yuan, Y.; Deng, Z.; Liu, T.; Dickschat, J. S. Angew. Chem., Int. Ed. 2018, 57, 15887–15890. doi:10.1002/anie.201809954
Return to citation in text: [1] -
Yang, Y.-l.; Zhang, S.; Ma, K.; Xu, Y.; Tao, Q.; Chen, Y.; Chen, J.; Guo, S.; Ren, J.; Wang, W.; Tao, Y.; Yin, W.-B.; Liu, H. Angew. Chem., Int. Ed. 2017, 56, 4749–4752. doi:10.1002/anie.201700565
Return to citation in text: [1] [2] -
Köksal, M.; Jin, Y.; Coates, R. M.; Croteau, R.; Christianson, D. W. Nature 2011, 469, 116–120. doi:10.1038/nature09628
Return to citation in text: [1] -
Rabe, P.; Rinkel, J.; Nubbemeyer, B.; Köllner, T. G.; Chen, F.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 15420–15423. doi:10.1002/anie.201608971
Return to citation in text: [1] -
Peng, B.; Plan, M. R.; Chrysanthopoulos, P.; Hodson, M. P.; Nielsen, L. K.; Vickers, C. E. Metab. Eng. 2017, 39, 209–219. doi:10.1016/j.ymben.2016.12.003
Return to citation in text: [1] -
Matsuda, Y.; Mitsuhashi, T.; Quan, Z.; Abe, I. Org. Lett. 2015, 17, 4644–4647. doi:10.1021/acs.orglett.5b02404
Return to citation in text: [1] -
Kim, D.; Lee, E.; Lee, J.; Leutou, A.; Shin, Y.-H.; Choi, B.; Hwang, J.; Hahn, D.; Choi, H.; Chin, J.; Cho, S.; Hong, Y.; Ko, J.; Seong, C.; Maloney, K.; Oh, D.-C.; Yang, I.; Hwang, H.; Nam, S.-J. Mar. Drugs 2018, 16, 130. doi:10.3390/md16040130
Return to citation in text: [1] -
Klapschinski, T. A.; Rabe, P.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 10141–10144. doi:10.1002/anie.201605425
Return to citation in text: [1] -
Ye, Y.; Minami, A.; Mandi, A.; Liu, C.; Taniguchi, T.; Kuzuyama, T.; Monde, K.; Gomi, K.; Oikawa, H. J. Am. Chem. Soc. 2015, 137, 11846–11853. doi:10.1021/jacs.5b08319
Return to citation in text: [1] -
Nakano, C.; Kudo, F.; Eguchi, T.; Ohnishi, Y. ChemBioChem 2011, 12, 2271–2275. doi:10.1002/cbic.201100418
Return to citation in text: [1] -
Hu, Y.; Chou, W. K. W.; Hopson, R.; Cane, D. E. Chem. Biol. 2011, 18, 32–37. doi:10.1016/j.chembiol.2010.11.008
Return to citation in text: [1] -
Cao, L.; Shehla, N.; Tasneem, S.; Cao, M.; Sheng, W.; Jian, Y.; Li, B.; Peng, C.; Choudhary, M. I.; Atta-ur-Rahman; Liao, D.-f.; Wang, W. Molecules 2019, 24, 1664. doi:10.3390/molecules24091664
Return to citation in text: [1] -
Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S. E.; Tang, Y. Nature 2018, 559, 415–418. doi:10.1038/s41586-018-0319-4
Return to citation in text: [1] -
Minami, A.; Ozaki, T.; Liu, C.; Oikawa, H. Nat. Prod. Rep. 2018, 35, 1330–1346. doi:10.1039/c8np00026c
Return to citation in text: [1] -
Bian, G.; Hou, A.; Yuan, Y.; Hu, B.; Cheng, S.; Ye, Z.; Di, Y.; Deng, Z.; Liu, T. Org. Lett. 2018, 20, 1626–1629. doi:10.1021/acs.orglett.8b00366
Return to citation in text: [1] [2] [3] [4] -
Mannina, L.; Segre, A. L.; Ritieni, A.; Fogliano, V.; Vinale, F.; Randazzo, G.; Maddau, L.; Bottalico, A. Tetrahedron 1997, 53, 3135–3144. doi:10.1016/s0040-4020(97)00024-0
Return to citation in text: [1] -
John, R. P.; Tyagi, R. D.; Prévost, D.; Brar, S. K.; Pouleur, S.; Surampalli, R. Y. Crop Prot. 2010, 29, 1452–1459. doi:10.1016/j.cropro.2010.08.004
Return to citation in text: [1] -
Bian, G.; Deng, Z.; Liu, T. Curr. Opin. Biotechnol. 2017, 48, 234–241. doi:10.1016/j.copbio.2017.07.002
Return to citation in text: [1] -
Peng, B.; Nielsen, L. K.; Kampranis, S. C.; Vickers, C. E. Metab. Eng. 2018, 47, 83–93. doi:10.1016/j.ymben.2018.02.005
Return to citation in text: [1] -
Yap, H.-Y. Y.; Muria-Gonzalez, M. J.; Kong, B.-H.; Stubbs, K. A.; Tan, C.-S.; Ng, S.-T.; Tan, N.-H.; Solomon, P. S.; Fung, S.-Y.; Chooi, Y.-H. Microb. Cell Fact. 2017, 16, 103. doi:10.1186/s12934-017-0713-x
Return to citation in text: [1] -
Kubicek, C. P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D. A.; Druzhinina, I. S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B. A.; Mukherjee, P. K.; Mukherjee, M.; Kredics, L.; Alcaraz, L. D.; Aerts, A.; Antal, Z.; Atanasova, L.; Cervantes-Badillo, M. G.; Challacombe, J.; Chertkov, O.; McCluskey, K.; Coulpier, F.; Deshpande, N.; von Döhren, H.; Ebbole, D. J.; Esquivel-Naranjo, E. U.; Fekete, E.; Flipphi, M.; Glaser, F.; Gómez-Rodríguez, E. Y.; Gruber, S.; Han, C.; Henrissat, B.; Hermosa, R.; Hernández-Oñate, M.; Karaffa, L.; Kosti, I.; Le Crom, S.; Lindquist, E.; Lucas, S.; Lübeck, M.; Lübeck, P. S.; Margeot, A.; Metz, B.; Misra, M.; Nevalainen, H.; Omann, M.; Packer, N.; Perrone, G.; Uresti-Rivera, E. E.; Salamov, A.; Schmoll, M.; Seiboth, B.; Shapiro, H.; Sukno, S.; Tamayo-Ramos, J. A.; Tisch, D.; Wiest, A.; Wilkinson, H. H.; Zhang, M.; Coutinho, P. M.; Kenerley, C. M.; Monte, E.; Baker, S. E.; Grigoriev, I. V. Genome Biol. 2011, 12, R40. doi:10.1186/gb-2011-12-4-r40
Return to citation in text: [1] -
Martinez, D.; Berka, R. M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S. E.; Chapman, J.; Chertkov, O.; Coutinho, P. M.; Cullen, D.; Danchin, E. G. J.; Grigoriev, I. V.; Harris, P.; Jackson, M.; Kubicek, C. P.; Han, C. S.; Ho, I.; Larrondo, L. F.; de Leon, A. L.; Magnuson, J. K.; Merino, S.; Misra, M.; Nelson, B.; Putnam, N.; Robbertse, B.; Salamov, A. A.; Schmoll, M.; Terry, A.; Thayer, N.; Westerholm-Parvinen, A.; Schoch, C. L.; Yao, J.; Barabote, R.; Nelson, M. A.; Detter, C.; Bruce, D.; Kuske, C. R.; Xie, G.; Richardson, P.; Rokhsar, D. S.; Lucas, S. M.; Rubin, E. M.; Dunn-Coleman, N.; Ward, M.; Brettin, T. S. Nat. Biotechnol. 2008, 26, 553–560. doi:10.1038/nbt1403
Return to citation in text: [1] -
Yuan, Y.; Litzenburger, M.; Cheng, S.; Bian, G.; Hu, B.; Yan, P.; Cai, Y.; Deng, Z.; Bernhardt, R.; Liu, T. ChemBioChem 2019, 20, 677–682. doi:10.1002/cbic.201800670
Return to citation in text: [1] -
Burkhardt, I.; Siemon, T.; Henrot, M.; Studt, L.; Rösler, S.; Tudzynski, B.; Christmann, M.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 8748–8751. doi:10.1002/anie.201603782
Return to citation in text: [1] [2] -
Burkhardt, I.; Kreuzenbeck, N. B.; Beemelmanns, C.; Dickschat, J. S. Org. Biomol. Chem. 2019, 17, 3348–3355. doi:10.1039/c8ob02744g
Return to citation in text: [1] -
Gallo, A.; Mulè, G.; Favilla, M.; Altomare, C. Physiol. Mol. Plant Pathol. 2004, 65, 11–20. doi:10.1016/j.pmpp.2004.11.005
Return to citation in text: [1] -
Crutcher, F. K.; Parich, A.; Schuhmacher, R.; Mukherjee, P. K.; Zeilinger, S.; Kenerley, C. M. Fungal Genet. Biol. 2013, 56, 67–77. doi:10.1016/j.fgb.2013.05.003
Return to citation in text: [1] -
Zhu, F.; Zhong, X.; Hu, M.; Lu, L.; Deng, Z.; Liu, T. Biotechnol. Bioeng. 2014, 111, 1396–1405. doi:10.1002/bit.25198
Return to citation in text: [1] -
Hu, D.-B.; Zhang, S.; He, J.-B.; Dong, Z.-J.; Li, Z.-H.; Feng, T.; Liu, J.-K. Fitoterapia 2015, 104, 50–54. doi:10.1016/j.fitote.2015.05.009
Return to citation in text: [1] [2] -
Iliopoulou, D.; Vagias, C.; Galanakis, D.; Argyropoulos, D.; Roussis, V. Org. Lett. 2002, 4, 3263–3266. doi:10.1021/ol026506z
Return to citation in text: [1] -
Takino, J.; Kozaki, T.; Sato, Y.; Liu, C.; Ozaki, T.; Minami, A.; Oikawa, H. J. Am. Chem. Soc. 2018, 140, 12392–12395. doi:10.1021/jacs.8b08925
Return to citation in text: [1] -
Wawrzyn, G. T.; Quin, M. B.; Choudhary, S.; López-Gallego, F.; Schmidt-Dannert, C. Chem. Biol. 2012, 19, 772–783. doi:10.1016/j.chembiol.2012.05.012
Return to citation in text: [1]
| 36. | Iliopoulou, D.; Vagias, C.; Galanakis, D.; Argyropoulos, D.; Roussis, V. Org. Lett. 2002, 4, 3263–3266. doi:10.1021/ol026506z |
| 8. | Yang, Y.-l.; Zhang, S.; Ma, K.; Xu, Y.; Tao, Q.; Chen, Y.; Chen, J.; Guo, S.; Ren, J.; Wang, W.; Tao, Y.; Yin, W.-B.; Liu, H. Angew. Chem., Int. Ed. 2017, 56, 4749–4752. doi:10.1002/anie.201700565 |
| 37. | Takino, J.; Kozaki, T.; Sato, Y.; Liu, C.; Ozaki, T.; Minami, A.; Oikawa, H. J. Am. Chem. Soc. 2018, 140, 12392–12395. doi:10.1021/jacs.8b08925 |
| 21. | Bian, G.; Hou, A.; Yuan, Y.; Hu, B.; Cheng, S.; Ye, Z.; Di, Y.; Deng, Z.; Liu, T. Org. Lett. 2018, 20, 1626–1629. doi:10.1021/acs.orglett.8b00366 |
| 38. | Wawrzyn, G. T.; Quin, M. B.; Choudhary, S.; López-Gallego, F.; Schmidt-Dannert, C. Chem. Biol. 2012, 19, 772–783. doi:10.1016/j.chembiol.2012.05.012 |
| 1. | Bian, G. K.; Ma, T.; Liu, T. G. Methods Enzymol. 2018, 608, 97–129. doi:10.1016/bs.mie.2018.04.025 |
| 2. | Huber, T.; Weisheit, L.; Magauer, T. Beilstein J. Org. Chem. 2015, 11, 2521–2539. doi:10.3762/bjoc.11.273 |
| 8. | Yang, Y.-l.; Zhang, S.; Ma, K.; Xu, Y.; Tao, Q.; Chen, Y.; Chen, J.; Guo, S.; Ren, J.; Wang, W.; Tao, Y.; Yin, W.-B.; Liu, H. Angew. Chem., Int. Ed. 2017, 56, 4749–4752. doi:10.1002/anie.201700565 |
| 21. | Bian, G.; Hou, A.; Yuan, Y.; Hu, B.; Cheng, S.; Ye, Z.; Di, Y.; Deng, Z.; Liu, T. Org. Lett. 2018, 20, 1626–1629. doi:10.1021/acs.orglett.8b00366 |
| 7. | Bian, G.; Rinkel, J.; Wang, Z.; Lauterbach, L.; Hou, A.; Yuan, Y.; Deng, Z.; Liu, T.; Dickschat, J. S. Angew. Chem., Int. Ed. 2018, 57, 15887–15890. doi:10.1002/anie.201809954 |
| 22. | Mannina, L.; Segre, A. L.; Ritieni, A.; Fogliano, V.; Vinale, F.; Randazzo, G.; Maddau, L.; Bottalico, A. Tetrahedron 1997, 53, 3135–3144. doi:10.1016/s0040-4020(97)00024-0 |
| 23. | John, R. P.; Tyagi, R. D.; Prévost, D.; Brar, S. K.; Pouleur, S.; Surampalli, R. Y. Crop Prot. 2010, 29, 1452–1459. doi:10.1016/j.cropro.2010.08.004 |
| 6. | Huang, A. C.; Hong, Y. J.; Bond, A. D.; Tantillo, D. J.; Osbourn, A. Angew. Chem., Int. Ed. 2018, 57, 1291–1295. doi:10.1002/anie.201711444 |
| 19. | Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S. E.; Tang, Y. Nature 2018, 559, 415–418. doi:10.1038/s41586-018-0319-4 |
| 3. | Christianson, D. W. Chem. Rev. 2017, 117, 11570–11648. doi:10.1021/acs.chemrev.7b00287 |
| 4. | Pemberton, T. A.; Chen, M.; Harris, G. G.; Chou, W. K. W.; Duan, L.; Köksal, M.; Genshaft, A. S.; Cane, D. E.; Christianson, D. W. Biochemistry 2017, 56, 2010–2023. doi:10.1021/acs.biochem.7b00137 |
| 5. | Lauterbach, L.; Rinkel, J.; Dickschat, J. S. Angew. Chem., Int. Ed. 2018, 57, 8280–8283. doi:10.1002/anie.201803800 |
| 20. | Minami, A.; Ozaki, T.; Liu, C.; Oikawa, H. Nat. Prod. Rep. 2018, 35, 1330–1346. doi:10.1039/c8np00026c |
| 13. | Kim, D.; Lee, E.; Lee, J.; Leutou, A.; Shin, Y.-H.; Choi, B.; Hwang, J.; Hahn, D.; Choi, H.; Chin, J.; Cho, S.; Hong, Y.; Ko, J.; Seong, C.; Maloney, K.; Oh, D.-C.; Yang, I.; Hwang, H.; Nam, S.-J. Mar. Drugs 2018, 16, 130. doi:10.3390/md16040130 |
| 15. | Ye, Y.; Minami, A.; Mandi, A.; Liu, C.; Taniguchi, T.; Kuzuyama, T.; Monde, K.; Gomi, K.; Oikawa, H. J. Am. Chem. Soc. 2015, 137, 11846–11853. doi:10.1021/jacs.5b08319 |
| 16. | Nakano, C.; Kudo, F.; Eguchi, T.; Ohnishi, Y. ChemBioChem 2011, 12, 2271–2275. doi:10.1002/cbic.201100418 |
| 17. | Hu, Y.; Chou, W. K. W.; Hopson, R.; Cane, D. E. Chem. Biol. 2011, 18, 32–37. doi:10.1016/j.chembiol.2010.11.008 |
| 3. | Christianson, D. W. Chem. Rev. 2017, 117, 11570–11648. doi:10.1021/acs.chemrev.7b00287 |
| 12. | Matsuda, Y.; Mitsuhashi, T.; Quan, Z.; Abe, I. Org. Lett. 2015, 17, 4644–4647. doi:10.1021/acs.orglett.5b02404 |
| 18. | Cao, L.; Shehla, N.; Tasneem, S.; Cao, M.; Sheng, W.; Jian, Y.; Li, B.; Peng, C.; Choudhary, M. I.; Atta-ur-Rahman; Liao, D.-f.; Wang, W. Molecules 2019, 24, 1664. doi:10.3390/molecules24091664 |
| 11. | Peng, B.; Plan, M. R.; Chrysanthopoulos, P.; Hodson, M. P.; Nielsen, L. K.; Vickers, C. E. Metab. Eng. 2017, 39, 209–219. doi:10.1016/j.ymben.2016.12.003 |
| 9. | Köksal, M.; Jin, Y.; Coates, R. M.; Croteau, R.; Christianson, D. W. Nature 2011, 469, 116–120. doi:10.1038/nature09628 |
| 10. | Rabe, P.; Rinkel, J.; Nubbemeyer, B.; Köllner, T. G.; Chen, F.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 15420–15423. doi:10.1002/anie.201608971 |
| 14. | Klapschinski, T. A.; Rabe, P.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 10141–10144. doi:10.1002/anie.201605425 |
| 27. | Kubicek, C. P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D. A.; Druzhinina, I. S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B. A.; Mukherjee, P. K.; Mukherjee, M.; Kredics, L.; Alcaraz, L. D.; Aerts, A.; Antal, Z.; Atanasova, L.; Cervantes-Badillo, M. G.; Challacombe, J.; Chertkov, O.; McCluskey, K.; Coulpier, F.; Deshpande, N.; von Döhren, H.; Ebbole, D. J.; Esquivel-Naranjo, E. U.; Fekete, E.; Flipphi, M.; Glaser, F.; Gómez-Rodríguez, E. Y.; Gruber, S.; Han, C.; Henrissat, B.; Hermosa, R.; Hernández-Oñate, M.; Karaffa, L.; Kosti, I.; Le Crom, S.; Lindquist, E.; Lucas, S.; Lübeck, M.; Lübeck, P. S.; Margeot, A.; Metz, B.; Misra, M.; Nevalainen, H.; Omann, M.; Packer, N.; Perrone, G.; Uresti-Rivera, E. E.; Salamov, A.; Schmoll, M.; Seiboth, B.; Shapiro, H.; Sukno, S.; Tamayo-Ramos, J. A.; Tisch, D.; Wiest, A.; Wilkinson, H. H.; Zhang, M.; Coutinho, P. M.; Kenerley, C. M.; Monte, E.; Baker, S. E.; Grigoriev, I. V. Genome Biol. 2011, 12, R40. doi:10.1186/gb-2011-12-4-r40 |
| 21. | Bian, G.; Hou, A.; Yuan, Y.; Hu, B.; Cheng, S.; Ye, Z.; Di, Y.; Deng, Z.; Liu, T. Org. Lett. 2018, 20, 1626–1629. doi:10.1021/acs.orglett.8b00366 |
| 24. | Bian, G.; Deng, Z.; Liu, T. Curr. Opin. Biotechnol. 2017, 48, 234–241. doi:10.1016/j.copbio.2017.07.002 |
| 25. | Peng, B.; Nielsen, L. K.; Kampranis, S. C.; Vickers, C. E. Metab. Eng. 2018, 47, 83–93. doi:10.1016/j.ymben.2018.02.005 |
| 26. | Yap, H.-Y. Y.; Muria-Gonzalez, M. J.; Kong, B.-H.; Stubbs, K. A.; Tan, C.-S.; Ng, S.-T.; Tan, N.-H.; Solomon, P. S.; Fung, S.-Y.; Chooi, Y.-H. Microb. Cell Fact. 2017, 16, 103. doi:10.1186/s12934-017-0713-x |
| 35. | Hu, D.-B.; Zhang, S.; He, J.-B.; Dong, Z.-J.; Li, Z.-H.; Feng, T.; Liu, J.-K. Fitoterapia 2015, 104, 50–54. doi:10.1016/j.fitote.2015.05.009 |
| 35. | Hu, D.-B.; Zhang, S.; He, J.-B.; Dong, Z.-J.; Li, Z.-H.; Feng, T.; Liu, J.-K. Fitoterapia 2015, 104, 50–54. doi:10.1016/j.fitote.2015.05.009 |
| 30. | Burkhardt, I.; Siemon, T.; Henrot, M.; Studt, L.; Rösler, S.; Tudzynski, B.; Christmann, M.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 8748–8751. doi:10.1002/anie.201603782 |
| 34. | Zhu, F.; Zhong, X.; Hu, M.; Lu, L.; Deng, Z.; Liu, T. Biotechnol. Bioeng. 2014, 111, 1396–1405. doi:10.1002/bit.25198 |
| 21. | Bian, G.; Hou, A.; Yuan, Y.; Hu, B.; Cheng, S.; Ye, Z.; Di, Y.; Deng, Z.; Liu, T. Org. Lett. 2018, 20, 1626–1629. doi:10.1021/acs.orglett.8b00366 |
| 32. | Gallo, A.; Mulè, G.; Favilla, M.; Altomare, C. Physiol. Mol. Plant Pathol. 2004, 65, 11–20. doi:10.1016/j.pmpp.2004.11.005 |
| 33. | Crutcher, F. K.; Parich, A.; Schuhmacher, R.; Mukherjee, P. K.; Zeilinger, S.; Kenerley, C. M. Fungal Genet. Biol. 2013, 56, 67–77. doi:10.1016/j.fgb.2013.05.003 |
| 28. | Martinez, D.; Berka, R. M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S. E.; Chapman, J.; Chertkov, O.; Coutinho, P. M.; Cullen, D.; Danchin, E. G. J.; Grigoriev, I. V.; Harris, P.; Jackson, M.; Kubicek, C. P.; Han, C. S.; Ho, I.; Larrondo, L. F.; de Leon, A. L.; Magnuson, J. K.; Merino, S.; Misra, M.; Nelson, B.; Putnam, N.; Robbertse, B.; Salamov, A. A.; Schmoll, M.; Terry, A.; Thayer, N.; Westerholm-Parvinen, A.; Schoch, C. L.; Yao, J.; Barabote, R.; Nelson, M. A.; Detter, C.; Bruce, D.; Kuske, C. R.; Xie, G.; Richardson, P.; Rokhsar, D. S.; Lucas, S. M.; Rubin, E. M.; Dunn-Coleman, N.; Ward, M.; Brettin, T. S. Nat. Biotechnol. 2008, 26, 553–560. doi:10.1038/nbt1403 |
| 29. | Yuan, Y.; Litzenburger, M.; Cheng, S.; Bian, G.; Hu, B.; Yan, P.; Cai, Y.; Deng, Z.; Bernhardt, R.; Liu, T. ChemBioChem 2019, 20, 677–682. doi:10.1002/cbic.201800670 |
| 30. | Burkhardt, I.; Siemon, T.; Henrot, M.; Studt, L.; Rösler, S.; Tudzynski, B.; Christmann, M.; Dickschat, J. S. Angew. Chem., Int. Ed. 2016, 55, 8748–8751. doi:10.1002/anie.201603782 |
| 31. | Burkhardt, I.; Kreuzenbeck, N. B.; Beemelmanns, C.; Dickschat, J. S. Org. Biomol. Chem. 2019, 17, 3348–3355. doi:10.1039/c8ob02744g |
© 2019 Sun et al.; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0). Please note that the reuse, redistribution and reproduction in particular requires that the authors and source are credited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (https://www.beilstein-journals.org/bjoc)