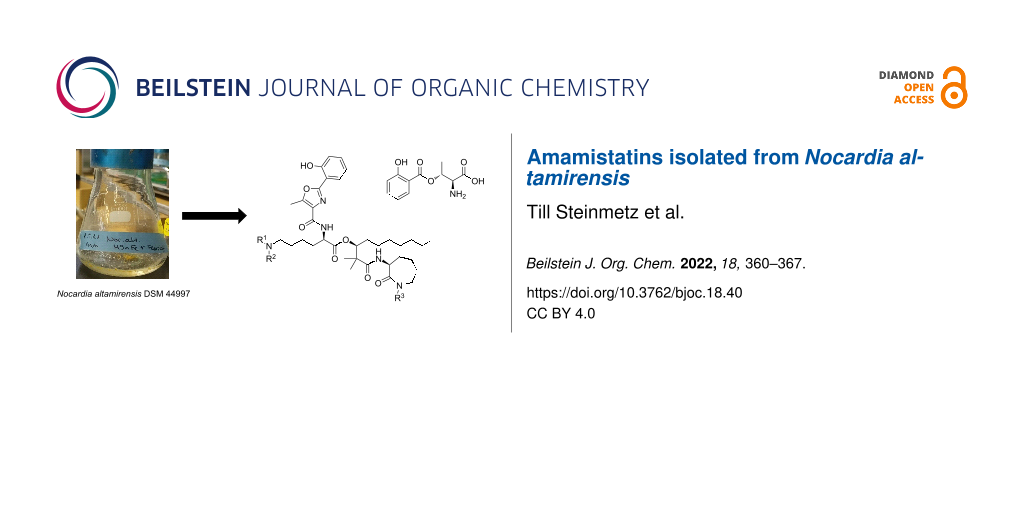
Abstract
Four new phenolic siderophores were isolated from the actinomycete Nocardia altamirensis along with the known natural product amamistatin B and a putative biosynthetic shunt product. The structures of all compounds were elucidated through 1D and 2D NMR analyses as well as mass spectrometry. The iron-chelating properties of the retrieved metabolites were evaluated in a chrome azurol S assay.
Graphical Abstract
Introduction
Iron is known to easily interconvert between a reduced ferrous (Fe2+) and an oxidized ferric state (Fe3+). This feature makes iron very useful as an enzyme cofactor for the shuffling of electrons. As a consequence of this, the transition metal is involved in many fundamental biological processes, such as respiration, photosynthesis, or nitrogen fixation [1]. In order to achieve iron homeostasis, organisms must be able to control the uptake of this important nutrient from the environment. In bacteria and fungi this is usually accomplished with the help of siderophores [2]. Siderophores are small molecules which, upon secretion, solubilize and coordinate ferric iron with high affinity. The ability to bind metallic ions depends on the presence of suitable ligand groups. Hydroxamates, 2,3-dihydroxybenzoates, and α-hydroxycarboxylates make excellent bidentate ligand groups, as their negatively charged oxygen atoms can maintain strong interactions with ferric iron [3]. But there are also many siderophores featuring ligand groups with nitrogen or sulfur as donor atoms [4]. The siderophore–iron complexes are recognized by highly selective microbial transporters. Following their translocation into the cell, the bound iron is released via a reductive or hydrolytic mechanism [2]
Members of the genus Nocardia are filamentous Actinobacteria, which live as saprophytes and were reported from a variety of environments. Although the soil is likely the primary reservoir of these bacteria, a considerable number of Nocardia strains was isolated from human tissue and can be associated with infectious diseases [5]. The genus Nocardia as a whole is considered to be a rich source of bioactive secondary metabolites, including many structurally diverse siderophores [6]. Up to now, however, chemical studies in this taxonomic group mainly focused on pathogenic strains. In the present study we explored siderophore production in the bacterium Nocardia altamirensis DSM 44997, which had originally been isolated from a microbial cave wall community and represents a non-pathogenic strain [5]. Our analysis led to the identification of six iron-chelating molecules belonging to the amamistatin family. Four of these compounds (1–4) have not been reported before. The isolated metabolite 5 represents the previously described siderophore amamistatin B [7], while compound 6 was before only known as a decomposition product of a synthetically prepared obafluorin derivative [8].
Results and Discussion
To induce siderophore biosynthesis in N. altamirensis DSM 44997, the bacterium was grown in minimal medium without the addition of iron salts. Initial evidence for siderophore production was obtained from a chrome azurol S (CAS) assay [9], which showed a clear color change from blue to pink upon testing of the culture broth. Several batch fermentations of N. altamirensis DSM 44997 were then carried out to secure sufficient material of the responsible iron chelator(s) for isolation and structure elucidation. The metabolites secreted into the culture broth were recovered post fermentation with the adsorber resin XAD-7. After removal of the culture supernatant by filtration, the adsorbed compounds were eluted from the resin with methanol. The resulting extract was concentrated to dryness and a first preliminary fractionation was accomplished by flash column chromatography on reversed-phase silica gel using increasing concentrations of methanol in water as eluent. Fractions that showed a color change in the CAS assay were pooled and subjected to semipreparative reversed-phase HPLC. This led to the isolation of six CAS active compounds (1–6; Figure 1).
Figure 1: Chemical structures of amamistatins (1–5) and a putative biosynthetic shunt product (6) isolated in this study. 1: R1 = CHO, R2 = H, R3 = H; 2: R1 = CHO, R2 = H, R3 = OH; 3: R1 = H, R2 = H, R3 = H; 4: R1 = H, R2 = OH, R3 = H; 5: R1 = CHO, R2 = OH, R3 = OH.
Figure 1: Chemical structures of amamistatins (1–5) and a putative biosynthetic shunt product (6) isolated in...
The major metabolite 1 (12 mg) was obtained as a slight reddish oil. High resolution (HR) ESIMS measurement revealed a molecular ion at m/z 684.3969 [M + H]+, which suggests a molecular formula of C36H53N5O8 (calcd for C36H54N5O8, 684.3972) and corresponds with 13 degrees of unsaturation. An inspection of the 13C NMR spectrum confirmed the number of carbon atoms derived from the MS measurement (Table 1).
Table 1: 1H and 13C NMR spectroscopic data for amamistatins (1–4) in methanol-d4.
| Pos. | 1 (amamistatin C) | 2 (amamistatin D) | 3 (amamistatin E) | 4 (amamistatin F) | ||||
|---|---|---|---|---|---|---|---|---|
| δH, M (J in Hz) | δC | δH, M (J in Hz) | δC | δH, M (J in Hz) | δC | δH, M (J in Hz) | δC | |
| α-amino-ε-caprolactam | ||||||||
| 1 | 176.9 | 170.7 | 177.1 | 177.1 | ||||
| 2 | 4.46 dd (1.6, 11.3) | 53.6 | 4.54 dd (1.3, 11.6) | 52.9 | 4.51 m | 52.0 | 4.49 m | 53.6 |
| 3 | 1.45 m, 1.89 m | 32.0 | 1.56 m, 1.92 m | 31.8 | 1.46 m, 1.91 m | 32.0 | 1.48 m, 1.57 m | 32.0 |
| 4 | 1.90 m, 1.77 m | 29.0 | 1.79 m | 28.7 | 1.92 m, 1.77 m | 29.0 | 1.92 m | 29.0 |
| 5 | 1.30 m | 30.3 | 1.80 m, 1.62 m | 27.2 | 1.31 m | 30.3 | 1.78 m, 1.29 m | 30.3 |
| 6 | 3.22 m | 42.6 |
3.93 dd (13.8, 16.0),
3.67 dd (5.0, 16.0) |
54.2 | 3.27 m | 42.6 | 3.28 m, 3.22 m | 42.6 |
| 2,2-dimethyl-3-hydroxydecanoic acid | ||||||||
| 7 | 176.5 | 176.8 | 176.6 | 176.5 | ||||
| 8 | 47.4 | 47.5 | 47.4 | 47.4 | ||||
| 9 | 5.09 dd (2.5, 10.4) | 80.3 | 5.17 dd (2.3, 10.3) | 80.3 | 5.14 dd (2.4, 10.5) | 80.5 | 5.15 dd (2.3, 10.5) | 80.5 |
| 10 | 1.57 m, 1.49 m | 31.0 | 1.59 m, 1.53 m | 31.0 | 1.58 m, 1.49 m | 30.9 | 1.57 m, 1.49 m | 30.9 |
| 11 | 1.25 m | 27.0 | 1.31 m | 26.9 | 1.25 m | 27.0 | 1.25 m | 27.1 |
| 12 | 1.19 m | 30.2 | 1.25 m | 30.3 | 1.21 m | 30.2 | 1.21 m | 30.2 |
| 13 | 1.28 m | 29.8 | 1.24 m | 30.4 | 1.31 m | 29.6 | 1.31 m | 30.3 |
| 14 | 1.20 m | 32.9 | 1.24 m | 32.9 | 1.20 m | 32.9 | 1.24 m | 32.9 |
| 15 | 1.22 m | 23.7 | 1.19 m | 23.7 | 1.22 m | 23.7 | 1.26 m | 23.5 |
| 16 | 0.82 (6.8) | 14.4 | 0.86 t (6.8) | 14.4 | 0.86 t (6.8) | 14.4 | 0.85 t (6.8) | 14.3 |
| 17 | 1.15 s | 23.7 | 1.21 s | 22.9 | 1.18 s | 23.7 | 1.18 s | 23.7 |
| 18 | 1.15 s | 21.6 | 1.21 s | 21.7 | 1.18 s | 21.4 | 1.18 s | 21.4 |
| lysine | ||||||||
| 19 | 172.5 | 172.8 | 172.3 | 172.3 | ||||
| 20 | 4.62 dd (5.3, 9.9) | 54.2 | 4.66 dd (5.1, 10.0) | 54.0 | 4.68 m | 53.6 | 4.69 m | 53.7 |
| 21 | 2.03 m,1.90 m | 31.6 | 2.05 m, 1.97 m | 31.6 | 2.03 m, 1.91 m | 31.5 | 2.09 m, 1.94 m | 31.4 |
| 22 | 1.58 m, 1.49 m | 24.6 | 1.60 m, 1.51 m | 24.5 | 1.81 m, 1.54 m | 24.2 | 1.55 m | 24.1 |
| 23 | 1.60 m, 1.76 m | 29.7 | 1.62 m, 1.80 m | 29.8 | 1.94 m | 27.6 | 1.74 m | 27.8 |
| 24 | 3.22 m | 38.6 | 3.26 m | 38.6 | 3.86 m | 64.9 | 2.96 m | 40.5 |
| 25 | 8.00 s | 163.9 | 8.03 s | 163.9 | – | – | – | – |
| asteroidic acid | ||||||||
| 26 | 163.5 | 163.5 | 163.5 | 163.6 | ||||
| 27 | 129.7 | 129.8 | 129.7 | 129.7 | ||||
| 28 | 154.4 | 154.4 | 154.5 | 154.5 | ||||
| 29 | 2.69 s | 11.7 | 2.73 s | 11.7 | 2.74 s | 11.7 | 2.72 s | 11.7 |
| 30 | 159.8 | 159.8 | 159.9 | 159.9 | ||||
| 31 | 111.7 | 111.8 | 111.7 | 111.7 | ||||
| 32 | 158.1 | 158.0 | 158.0 | 158.0 | ||||
| 33 | 7.03 dd (1.2, 8.3) | 118.3 | 7.05 (0.9, 8.5) | 118.2 | 7.06 dd (1.2, 8.4) | 118.2 | 7.06 dd (1.2, 8.4) | 118.2 |
| 34 | 7.38 ddd (1.7, 7.3, 8.3) | 133.9 | 7.42 (1.6, 7.4, 8.5) | 133.9 | 7.42 ddd (1.8, 7.3, 8.4) | 134.0 | 7.43 ddd (1.7, 7.4, 8.4) | 134.0 |
| 35 | 6.97 ddd (1.2, 7.3, 7.9) | 120.9 | 7.02 (0.9, 7.4, 7.9) | 120.9 | 7.02 ddd (1.2, 7.3, 8.1) | 121.0 | 7.02 ddd (1.2, 7.4, 8.1) | 121.0 |
| 36 | 7.83 dd (1.7, 7.9) | 127.5 | 7.87 dd (1.6, 7.9) | 127.6 | 7.87 dd (1.8, 8.1) | 127.5 | 7.87 dd (1.7, 8.1) | 127.5 |
Furthermore, the observed chemical shifts indicated that 14 carbons must be sp2-hybridized. Six of them form carbon–heteroatom double bonds (δC 176.9, 176.5, 172.5, 163.9, 163.5, 159.8 ppm) and eight of them are engaged in carbon–carbon double bonds (δC 158.1, 154.4, 133.9, 129.7, 127.5, 120.9, 118.3, 111.7 ppm). Therefore, compound 1 must feature three ring structures in order to comply with the required degrees of unsaturation. Proton–proton correlation spectroscopy (COSY) revealed four spin systems, which could be connected through 1H,13C long-range correlations (Figure 2) determined in a heteronuclear multiple bond correlation (HMBC) experiment. The first spin system comprises the proton signals at δH 4.46 (dd, J = 11.3, 1.6 Hz, H-2), 3.22 (m, H2-6), 1.90 (m, H-4a), 1.77 (m, H-4b), 1.89 (m, H-3a), 1.45 (m, H-3b), and 1.30 ppm (m, H2-5). HMBC correlations from H-2 and H-6 to the carbonyl carbon C-1 in combination with the observed chemical shifts reinforced the assumption of an α-amino-ε-caprolactam structure. An HMBC correlation from H-2 to a carbonyl carbon at 176.5 ppm (C-7) allowed the linkage of this moiety with a 2,2-dimethyl-3-hydroxydecanoic acid residue, which was identified on the basis of 2D NMR data. In the HMBC spectrum, the singlet methyl protons H3-17 and H3-18 show correlations to C-7, a quaternary carbon at 47.4 ppm (C-8), and an oxymethine carbon at 80.3 ppm (C-9). COSY and HMBC data established the alkyl chain from CH-9 to CH3-16. A correlation between H-9 and C-19 in the HMBC spectrum unveiled the ester linkage with a lysine residue. The spin system of the latter includes proton resonances at δH 4.62 (dd, J = 9.9, 5.3 Hz, H-20), 2.03 ppm (m, H-21a), 1.90 ppm (m, H-21b), 1.58 (m, H-22a), 1.49 (m, H-22b), 1.76 (m, H-23a), 1.60 (m, H-23b), and 3.22 ppm (m, H2-24). HMBC correlations indicated the presence of a formyl group at the ε-amino group of this residue. The fourth spin system belongs to a 1,2-disubstituted benzene moiety, featuring proton signals at δH 7.03 (dd, J = 8.3, 1.2 Hz, H-33), 7.38 (ddd, J = 8.3, 7.3, 1.7 Hz, H-34), 6.97 (ddd, J = 7.9, 7.3, 1.2 Hz, H-35), and 7.83 ppm (dd, J = 7.9, 1.7 Hz, H-36). The upfield-shifted resonances of H-33 and H-35 suggested an electron-donating substituent at C-32 and the chemical shift of the latter (δC 158.1 ppm) supported the assignment of a hydroxy function in this position. From an analysis of the chemical shifts of the remaining carbon atoms (C-26 to C-30) as well as HMBC correlations, we concluded the presence of a 5-methyloxazole-4-carboxylate-derived residue adjacent to the benzene ring. This partial structure is a known motif in some siderophores and is referred to as asteroidic acid [7,10,11]. Due to an HMBC correlation from H-20 to C-26 the asteroidic acid moiety can be connected with the rest of the molecule, thereby completing the determination of the planar structure of 1.
Figure 2: 1H,1H COSY and selected 1H,13C HMBC correlations in compounds 1 and 6.
Figure 2: 1H,1H COSY and selected 1H,13C HMBC correlations in compounds 1 and 6.
Compound 2 (1.1 mg) was obtained as a clear oil. It possesses a molecular ion at m/z 700.3921 [M + H]+, which is consistent with a molecular formula of C36H53N5O9 (calcd for C36H54N5O9, 700.3922). The difference of 16 mass units in comparison to compound 1 suggests the presence of an additional oxygen atom. Comparison of the NMR data with 1 reveals the most significant chemical shift deviations in the α-amino-ε-caprolactam moiety. Since the splitting pattern of the corresponding proton signals is consistent with 1, the lactam nitrogen must be hydroxylated in 2. Tandem MS data is consistent with this assumption (Figure 3).
Figure 3: MS–MS fragmentation of amamistatins (1–5).
Figure 3: MS–MS fragmentation of amamistatins (1–5).
Compound 3 (1.8 mg) was isolated as a clear oil. Its molecular formula was established as C35H53N5O7 by the [M + H]+ ion at m/z 656.4019 (calcd for C35H54N5O7, 656.4023). Analysis of 1D and 2D NMR spectra confirmed that the aldehyde group in position 25 is missing. Therefore, compound 3 represents the N-desformyl analogue of 1.
Compound 4 (1.9 mg) was obtained as a reddish oil. Its molecular formula of C35H53N5O8 was determined by HRESIMS with a molecular ion at m/z 672.3966 [M + H]+ (calcd for C35H54N5O9, 672.3972). Similar to 3, compound 4 is also missing a formyl group at the ε-amino group of its lysine residue. However, its increased mass of 16 Da indicates the presence of an additional oxygen atom. A comparison of the NMR data with 3 revealed that the chemical shifts of CH2-24 are significantly shifted upfield. In contrast, all other resonances are highly conserved and no difference in the splitting pattern was observed. This suggests the presence of an electron-donating hydroxy group next to the ε-amino group in the lysine moiety. The assumption of a hydroxylamine function is consistent with tandem MS data, which showed mass shifts of 16 Da in the corresponding region in comparison to compound 3.
Compound 5 (0.7 mg) was obtained as a clear oil. Its molecular formula of C36H53N5O10 was determined by HRESIMS with a molecular ion at m/z 716.3820 [M + H]+ (calcd for C36H54N5O10, 716.3820). A comparison with published data led to the identification of compound 5 as the known siderophore amamistatin B [7].
Compound 6 (2.4 mg) was isolated as a slight reddish oil. The molecular formula of C11H13NO5 was derived from a molecular ion at m/z 240.0890 [M + H]+ (calcd for C11H14NO5, 240.0877). An inspection of the 13C NMR spectrum confirmed the calculated number of carbon atoms (Table 2). Furthermore, a total of eight sp2-hybridized carbons can be assigned based on their chemical shifts. Two of them are engaged in carbon–heteroatom double bonds (δC 170.0, 169.3 ppm) and six of them form carbon–carbon double bonds (δC 163.0, 137.5, 131.5, 120.5. 118.5, 113.0 ppm). This indicates the presence of one ring structure to comply with the required six degrees of unsaturation. With the help of COSY and HMBC data two residues were identified, including a threonine and a salicylic acid moiety. The two partial structures can be connected via an ester bond due to a key HMBC correlation from H-3 to C-5.
Table 2: 1H and 13C NMR spectroscopic data of isolated compound 6 in methanol-d4.
| Position | Compound 6 | |
|---|---|---|
| δC | δH, M (J in Hz) | |
| threonine | ||
| 1 | 169.3 | |
| 2 | 57.7 | 4.38 d (3.6 Hz) |
| 3 | 70.6 | 5.78 dq (3.6, 6.7 Hz) |
| 4 | 16.9 | 1.57 d (6.7 Hz) |
| salicylic acid | ||
| 5 | 170.0 | |
| 6 | 163.0 | |
| 7 | 113.0 | |
| 8 | 118.5 | 6.97 dd (1.0, 8.4 Hz) |
| 9 | 120.5 | 6.93 ddd (1.6, 7.2, 8.4 Hz) |
| 10 | 137.5 | 7.53 ddd (1.0, 7.2, 8.0 Hz) |
| 11 | 131.5 | 7.98 dd (1.6, 8.0 Hz) |
To analyze the configuration of the isolated compounds, their optical rotations were measured and compared with literature values of known natural products. The optical rotation of compound 5 was consistent with the reported value for amamistatin B ( = −9.8) [7]. Similar optical rotations were determined for 1, 3, and 4 (
= −1.4, −11.8, and −11.5, respectively). Furthermore, these compounds did not show any discrepancies in their NOESY spectra in comparison to 5. This suggests that 1, 3, 4, and 5 possess the same absolute configuration. Measurements of 2 were not possible due to the low quantity of isolated material, but it stands to reason that its configuration is identical to 5 under the assumption of a shared biosynthetic origin. Compound 6 shows an optical rotation of
= −10.6, which is consistent with the published value of pseudomonin A (
= −9.5) [12]. This natural product is structurally closely related to 6, featuring a terminal amide instead of a carboxylic acid function. Due to the similarity of both molecules, we propose that compound 6 possesses the same absolute configuration as pseudomonin A.
The affinities of compound 1–6 for the coordination of ferric iron were evaluated with the CAS assay [9] and compared with EDTA [13]. While the half-maximal displacement concentration (DC50) value of 5 (198 ± 10 μM) was comparable with EDTA (173 ± 6 μM), the other tested amamistatins were much weaker chelating agents (1, 3359 ± 181 μM; 2, 1074 ± 65 μM; 3, 3268 ± 183 μM; 4, 3298 ± 261 μM). The increase of the DC50 values (5 < 2 < 1 ≈ 3 ≈ 4) was in full accordance with the structural features of these molecules. Amamistatin B (5) exhibits three bidentate ligand groups and can thus form an octahedral complex with ferric iron. The removal of one of these groups (e.g., the hydroxamate function at the ε terminus of the lysine residue) already leads to a considerable loss of iron affinity, as observed for compound 2. This trend continues in 1, 3, and 4, each of which features only a single bidentate ligand group. Compound 6 possesses the least iron affinity with a DC50 value of 5008 ± 222 μM. The results show a clear correlation between the number of ligands in a molecule and its iron affinity.
On the basis of biosynthetic precedence [14], a plausible pathway for the natural products isolated in this study can be postulated (Figure 4). According to this proposal, compounds 1–4 are intermediates in the formation of 5, which has the strongest iron affinity and likely represents the end product of the pathway in N. altamirensis DSM 44997. Previous studies have demonstrated that, in nature, hydroxamates arise from the oxidation of terminal amino groups in amino acid side chains, followed by formylation or acylation of the resulting hydroxylamines. While the oxidations are catalyzed by flavin-dependent monooxygenases [12-16], the carbon transfer is mediated either by tetrahydrofolate-dependent formyl [17] or by acyl-CoA-dependent acyl transferases [13,15,16]. It is widely assumed that the oxidation precedes the formylation or acylation [17-22]. In this study, however, two molecules were isolated (1 and 3), the structures of which suggest that the timing of the enzymatic reactions can be interchanged. An alternative explanation would be that compound 5 is metabolized to 1, even though we did not find any literature evidence for such a reductive degradation of amamistatin-type siderophores.
Figure 4: Proposed biosynthesis of amamistatins isolated in this study.
Figure 4: Proposed biosynthesis of amamistatins isolated in this study.
Compound 6 represents a possible shunt product of amamistatin biosynthesis. A similar molecule and its putative biosynthetic pathway were recently described by Jaspars et al. [12]. In accordance with this proposal, salicylic acid and ʟ-threonine would form a salimethyloxazolinyl-thioester intermediate, which then undergoes a C–N bond opening resulting in an ester formation. Eventually, the thioester undergoes a hydrolysis reaction leading to compound 6.
The compounds, which were discovered in this study, are members of the large family of nocobactin-type siderophores. Structurally, compounds 1–5 are closely related to formobactin [10], brasilibactin [23], nocardimicin [24], and terpenibactins [15]. While the core scaffold of these natural products is conserved, they differ in the length of their fatty acid residues, their methylation pattern, and the presence of an oxazole or an oxazoline ring. Up to now, it is not clear whether these structural variations are biologically relevant in terms of iron sequestration and cellular uptake.
Conclusion
In summary, Nocardia altamirensis was found to synthesize various amamistatins under iron-deficient conditions. The structures of these natural products were verified by high-resolution mass spectrometry, tandem mass spectrometry as well as 1D and 2D NMR analyses. Their absolute configuration is consistent with the already known amamistatin B according to a comparison of optical rotation values and NOE data. The newly found compounds are assumed to represent intermediates or shunt products in amamistatin biosynthesis. In comparison to amamistatin B, they exhibit a lower iron affinity, which can be ascribed to the lack of hydroxamate groups.
Experimental
Analytical methods
LC–MS analysis was performed on a compact quadrupole-time of flight (Q-TOF) mass spectrometer from Bruker Daltonics with an Agilent 1260 Infinity LC system equipped with a Nucleoshell RP18 column (150 × 2.0 mm, Macherey-Nagel). NMR spectra were recorded on a Bruker 600 MHz Avance III HD system with methanol-d4 as solvent and internal standard. The solvent signal was referenced to δH 3.31 ppm and δC 49.0 ppm, respectively. Optical rotation was measured at 20 °C on a PerkinElmer polarimeter 341 with a sodium lamp (wavelength = 589 nm) using a 1 dm cuvette. For this, samples were dissolved in 1 mL methanol.
Cultivation and extraction of Nocardia altamirensis
For siderophore production, the strain was grown in 5-L Erlenmeyer flasks containing 1 L of iron-free M9 medium: 7.52 g/L Na2HPO4×2H2O, 3.0 g/L KH2PO4, 0.5 g/L NaCl, 0.5 g/L NH4Cl, 4 g/L glucose, 0.247 g/L MgSO4×7H2O, 0.074 g CaCl2×2H2O, 1 mL biotin (1 mg/mL), 1 mL thiamin·HCl (1 mg/mL), 10 mL trace elements solution (0.05 g/L EDTA, 0.84 mg/L ZnCl2, 0.13 mg/L CuCl2×2H2O, 0.01 mg/L CoCl2×2H2O, 0.01 mg/L H3BO3, 0.0016 mg/L MnCl2×4H2O). The cultivation was conducted on a rotary shaker at 130 rpm and 30 °C for three weeks. Afterwards, adsorber resin (XAD-7, 20 g/L) was added to the culture broth to bind the secreted metabolites. The resin was separated from the culture broth by filtration, washed with distilled water, and exhaustively extracted with methanol.
Isolation of metabolites 1–6
The concentrated extract was first fractionated by flash column chromatography over Polygoprep 60-50 C18 (Macherey-Nagel) using an increasing concentration of methanol in water. Fractions that gave a color change in the CAS assay were prepurified by reversed-phase HPLC using a Nucleodur C18 ec column (125 × 21 mm, 5 μm, Macherey-Nagel) and a gradient of methanol in water supplemented with 0.1% (v/v) trifluoroacetic acid. The gradient conditions were as follows: from 40% methanol to 70% in 5 minutes followed by an increase to 90% in 20 minutes, kept at 90% for 10 minutes. The flow rate was set to 10 mL/min. The elution of compounds was monitored with a diode array detector. Again, CAS active fractions were collected. For the final purification step, a Nucleodur C18 Isis column (250 × 10 mm, 5 μm, Macherey-Nagel) was used with a linear gradient from 25% to 90% acetonitrile in water + 0.1% (v/v) trifluoroacetic acid over a period of 30 min. The flow rate was set to 6 mL/min. The elution of compounds was monitored with a diode array detector.
Chrome azurol S (CAS) assay
The CAS assay solution was prepared as previously described [9]. This solution was aliquoted (100 μL) into the wells of a 96-well flat-bottomed microtiter plate. Subsequently, 5 μL test solution (= serial dilution of compounds 1–6 dissolved in DMSO) were added to every well. EDTA was used as a reference and dissolved in water. The microtiter plate was gently shaken for 4 h at room temperature. After this incubation period the remaining CAS–Fe3+ complex in each well was quantified by measuring the absorbance at 630 nm in a microplate reader. The values were plotted and the DC50 values were calculated. All tests were run in triplicate.
Supporting Information
| Supporting Information File 1: Copies of MS/MS and NMR spectra for compounds 1–6. | ||
| Format: PDF | Size: 2.1 MB | Download |
References
-
Hofmann, M.; Retamal-Morales, G.; Tischler, D. Nat. Prod. Rep. 2020, 37, 1262–1283. doi:10.1039/c9np00058e
Return to citation in text: [1] -
Raymond, K. N.; Allred, B. E.; Sia, A. K. Acc. Chem. Res. 2015, 48, 2496–2505. doi:10.1021/acs.accounts.5b00301
Return to citation in text: [1] [2] -
Kurth, C.; Kage, H.; Nett, M. Org. Biomol. Chem. 2016, 14, 8212–8227. doi:10.1039/c6ob01400c
Return to citation in text: [1] -
Johnstone, T. C.; Nolan, E. M. Dalton Trans. 2015, 44, 6320–6339. doi:10.1039/c4dt03559c
Return to citation in text: [1] -
Jurado, V.; Boiron, P.; Kroppenstedt, R. M.; Laurent, F.; Couble, A.; Laiz, L.; Klenk, H.-P.; Gonzalez, J. M.; Saiz-Jimenez, C.; Mouniee, D.; Bergeron, E.; Rodriguez-Nava, V. Int. J. Syst. Evol. Microbiol. 2008, 58, 2210–2214. doi:10.1099/ijs.0.65482-0
Return to citation in text: [1] [2] -
Engelbrecht, A.; Saad, H.; Gross, H.; Kaysser, L. Microb. Physiol. 2021, 31, 217–232. doi:10.1159/000516864
Return to citation in text: [1] -
Kokubo, S.; Suenaga, K.; Shinohara, C.; Tsuji, T.; Uemura, D. Tetrahedron 2000, 56, 6435–6440. doi:10.1016/s0040-4020(00)00591-3
Return to citation in text: [1] [2] [3] [4] -
Pu, Y.; Lowe, C.; Sailer, M.; Vederas, J. C. J. Org. Chem. 1994, 59, 3642–3655. doi:10.1021/jo00092a025
Return to citation in text: [1] -
Schwyn, B.; Neilands, J. B. Anal. Biochem. 1987, 160, 47–56. doi:10.1016/0003-2697(87)90612-9
Return to citation in text: [1] [2] [3] -
Murakami, Y.; Kato, S.; Nakajima, M.; Matsuoka, M.; Kawai, H.; Shin-Ya, K.; Seto, H. J. Antibiot. 1996, 49, 839–845. doi:10.7164/antibiotics.49.839
Return to citation in text: [1] [2] -
Hoshino, Y.; Chiba, K.; Ishino, K.; Fukai, T.; Igarashi, Y.; Yazawa, K.; Mikami, Y.; Ishikawa, J. J. Bacteriol. 2011, 193, 441–448. doi:10.1128/jb.00897-10
Return to citation in text: [1] -
Oluwabusola, E. T.; Adebisi, O. O.; Reyes, F.; Acquah, K. S.; de la Cruz, M.; Mweetwa, L. L.; Rajakulendran, J. E.; Warner, D. F.; Hai, D.; Ebel, R.; Jaspars, M. Beilstein J. Org. Chem. 2021, 17, 2390–2398. doi:10.3762/bjoc.17.156
Return to citation in text: [1] [2] [3] -
Igarashi, Y.; Ge, Y.; Zhou, T.; Sharma, A. R.; Harunari, E.; Oku, N.; Trianto, A. Beilstein J. Org. Chem. 2022, 18, 110–119. doi:10.3762/bjoc.18.12
Return to citation in text: [1] [2] [3] -
Chen, J.; Frediansyah, A.; Männle, D.; Straetener, J.; Brötz-Oesterhelt, H.; Ziemert, N.; Kaysser, L.; Gross, H. ChemBioChem 2020, 21, 2205–2213. doi:10.1002/cbic.202000062
Return to citation in text: [1] [2] -
Kreutzer, M. F.; Kage, H.; Nett, M. J. Am. Chem. Soc. 2012, 134, 5415–5422. doi:10.1021/ja300620z
Return to citation in text: [1] [2] [3] -
Kreutzer, M. F.; Nett, M. Org. Biomol. Chem. 2012, 10, 9338–9343. doi:10.1039/c2ob26296g
Return to citation in text: [1] [2] -
Heemstra, J. R., Jr.; Walsh, C. T.; Sattely, E. S. J. Am. Chem. Soc. 2009, 131, 15317–15329. doi:10.1021/ja9056008
Return to citation in text: [1] [2] -
Robbel, L.; Helmetag, V.; Knappe, T. A.; Marahiel, M. A. Biochemistry 2011, 50, 6073–6080. doi:10.1021/bi200699x
Return to citation in text: [1] -
Bosello, M.; Mielcarek, A.; Giessen, T. W.; Marahiel, M. A. Biochemistry 2012, 51, 3059–3066. doi:10.1021/bi201837f
Return to citation in text: [1] -
Lautru, S.; Deeth, R. J.; Bailey, L. M.; Challis, G. L. Nat. Chem. Biol. 2005, 1, 265–269. doi:10.1038/nchembio731
Return to citation in text: [1] -
Barona-Gómez, F.; Wong, U.; Giannakopulos, A. E.; Derrick, P. J.; Challis, G. L. J. Am. Chem. Soc. 2004, 126, 16282–16283. doi:10.1021/ja045774k
Return to citation in text: [1] -
Thariath, A.; Socha, D.; Valvano, M. A.; Viswanatha, T. J. Bacteriol. 1993, 175, 589–596. doi:10.1128/jb.175.3.589-596.1993
Return to citation in text: [1] -
Tsuda, M.; Yamakawa, M.; Oka, S.; Tanaka, Y.; Hoshino, Y.; Mikami, Y.; Sato, A.; Fujiwara, H.; Ohizumi, Y.; Kobayashi, J. J. Nat. Prod. 2005, 68, 462–464. doi:10.1021/np0496385
Return to citation in text: [1] -
Ikeda, Y.; Nonaka, H.; Furumai, T.; Onaka, H.; Igarashi, Y. J. Nat. Prod. 2005, 68, 1061–1065. doi:10.1021/np050091j
Return to citation in text: [1]
| 9. | Schwyn, B.; Neilands, J. B. Anal. Biochem. 1987, 160, 47–56. doi:10.1016/0003-2697(87)90612-9 |
| 1. | Hofmann, M.; Retamal-Morales, G.; Tischler, D. Nat. Prod. Rep. 2020, 37, 1262–1283. doi:10.1039/c9np00058e |
| 2. | Raymond, K. N.; Allred, B. E.; Sia, A. K. Acc. Chem. Res. 2015, 48, 2496–2505. doi:10.1021/acs.accounts.5b00301 |
| 12. | Oluwabusola, E. T.; Adebisi, O. O.; Reyes, F.; Acquah, K. S.; de la Cruz, M.; Mweetwa, L. L.; Rajakulendran, J. E.; Warner, D. F.; Hai, D.; Ebel, R.; Jaspars, M. Beilstein J. Org. Chem. 2021, 17, 2390–2398. doi:10.3762/bjoc.17.156 |
| 4. | Johnstone, T. C.; Nolan, E. M. Dalton Trans. 2015, 44, 6320–6339. doi:10.1039/c4dt03559c |
| 9. | Schwyn, B.; Neilands, J. B. Anal. Biochem. 1987, 160, 47–56. doi:10.1016/0003-2697(87)90612-9 |
| 3. | Kurth, C.; Kage, H.; Nett, M. Org. Biomol. Chem. 2016, 14, 8212–8227. doi:10.1039/c6ob01400c |
| 7. | Kokubo, S.; Suenaga, K.; Shinohara, C.; Tsuji, T.; Uemura, D. Tetrahedron 2000, 56, 6435–6440. doi:10.1016/s0040-4020(00)00591-3 |
| 2. | Raymond, K. N.; Allred, B. E.; Sia, A. K. Acc. Chem. Res. 2015, 48, 2496–2505. doi:10.1021/acs.accounts.5b00301 |
| 7. | Kokubo, S.; Suenaga, K.; Shinohara, C.; Tsuji, T.; Uemura, D. Tetrahedron 2000, 56, 6435–6440. doi:10.1016/s0040-4020(00)00591-3 |
| 7. | Kokubo, S.; Suenaga, K.; Shinohara, C.; Tsuji, T.; Uemura, D. Tetrahedron 2000, 56, 6435–6440. doi:10.1016/s0040-4020(00)00591-3 |
| 9. | Schwyn, B.; Neilands, J. B. Anal. Biochem. 1987, 160, 47–56. doi:10.1016/0003-2697(87)90612-9 |
| 5. | Jurado, V.; Boiron, P.; Kroppenstedt, R. M.; Laurent, F.; Couble, A.; Laiz, L.; Klenk, H.-P.; Gonzalez, J. M.; Saiz-Jimenez, C.; Mouniee, D.; Bergeron, E.; Rodriguez-Nava, V. Int. J. Syst. Evol. Microbiol. 2008, 58, 2210–2214. doi:10.1099/ijs.0.65482-0 |
| 7. | Kokubo, S.; Suenaga, K.; Shinohara, C.; Tsuji, T.; Uemura, D. Tetrahedron 2000, 56, 6435–6440. doi:10.1016/s0040-4020(00)00591-3 |
| 10. | Murakami, Y.; Kato, S.; Nakajima, M.; Matsuoka, M.; Kawai, H.; Shin-Ya, K.; Seto, H. J. Antibiot. 1996, 49, 839–845. doi:10.7164/antibiotics.49.839 |
| 11. | Hoshino, Y.; Chiba, K.; Ishino, K.; Fukai, T.; Igarashi, Y.; Yazawa, K.; Mikami, Y.; Ishikawa, J. J. Bacteriol. 2011, 193, 441–448. doi:10.1128/jb.00897-10 |
| 6. | Engelbrecht, A.; Saad, H.; Gross, H.; Kaysser, L. Microb. Physiol. 2021, 31, 217–232. doi:10.1159/000516864 |
| 5. | Jurado, V.; Boiron, P.; Kroppenstedt, R. M.; Laurent, F.; Couble, A.; Laiz, L.; Klenk, H.-P.; Gonzalez, J. M.; Saiz-Jimenez, C.; Mouniee, D.; Bergeron, E.; Rodriguez-Nava, V. Int. J. Syst. Evol. Microbiol. 2008, 58, 2210–2214. doi:10.1099/ijs.0.65482-0 |
| 8. | Pu, Y.; Lowe, C.; Sailer, M.; Vederas, J. C. J. Org. Chem. 1994, 59, 3642–3655. doi:10.1021/jo00092a025 |
| 12. | Oluwabusola, E. T.; Adebisi, O. O.; Reyes, F.; Acquah, K. S.; de la Cruz, M.; Mweetwa, L. L.; Rajakulendran, J. E.; Warner, D. F.; Hai, D.; Ebel, R.; Jaspars, M. Beilstein J. Org. Chem. 2021, 17, 2390–2398. doi:10.3762/bjoc.17.156 |
| 13. | Igarashi, Y.; Ge, Y.; Zhou, T.; Sharma, A. R.; Harunari, E.; Oku, N.; Trianto, A. Beilstein J. Org. Chem. 2022, 18, 110–119. doi:10.3762/bjoc.18.12 |
| 14. | Chen, J.; Frediansyah, A.; Männle, D.; Straetener, J.; Brötz-Oesterhelt, H.; Ziemert, N.; Kaysser, L.; Gross, H. ChemBioChem 2020, 21, 2205–2213. doi:10.1002/cbic.202000062 |
| 15. | Kreutzer, M. F.; Kage, H.; Nett, M. J. Am. Chem. Soc. 2012, 134, 5415–5422. doi:10.1021/ja300620z |
| 16. | Kreutzer, M. F.; Nett, M. Org. Biomol. Chem. 2012, 10, 9338–9343. doi:10.1039/c2ob26296g |
| 13. | Igarashi, Y.; Ge, Y.; Zhou, T.; Sharma, A. R.; Harunari, E.; Oku, N.; Trianto, A. Beilstein J. Org. Chem. 2022, 18, 110–119. doi:10.3762/bjoc.18.12 |
| 14. | Chen, J.; Frediansyah, A.; Männle, D.; Straetener, J.; Brötz-Oesterhelt, H.; Ziemert, N.; Kaysser, L.; Gross, H. ChemBioChem 2020, 21, 2205–2213. doi:10.1002/cbic.202000062 |
| 24. | Ikeda, Y.; Nonaka, H.; Furumai, T.; Onaka, H.; Igarashi, Y. J. Nat. Prod. 2005, 68, 1061–1065. doi:10.1021/np050091j |
| 15. | Kreutzer, M. F.; Kage, H.; Nett, M. J. Am. Chem. Soc. 2012, 134, 5415–5422. doi:10.1021/ja300620z |
| 10. | Murakami, Y.; Kato, S.; Nakajima, M.; Matsuoka, M.; Kawai, H.; Shin-Ya, K.; Seto, H. J. Antibiot. 1996, 49, 839–845. doi:10.7164/antibiotics.49.839 |
| 23. | Tsuda, M.; Yamakawa, M.; Oka, S.; Tanaka, Y.; Hoshino, Y.; Mikami, Y.; Sato, A.; Fujiwara, H.; Ohizumi, Y.; Kobayashi, J. J. Nat. Prod. 2005, 68, 462–464. doi:10.1021/np0496385 |
| 17. | Heemstra, J. R., Jr.; Walsh, C. T.; Sattely, E. S. J. Am. Chem. Soc. 2009, 131, 15317–15329. doi:10.1021/ja9056008 |
| 18. | Robbel, L.; Helmetag, V.; Knappe, T. A.; Marahiel, M. A. Biochemistry 2011, 50, 6073–6080. doi:10.1021/bi200699x |
| 19. | Bosello, M.; Mielcarek, A.; Giessen, T. W.; Marahiel, M. A. Biochemistry 2012, 51, 3059–3066. doi:10.1021/bi201837f |
| 20. | Lautru, S.; Deeth, R. J.; Bailey, L. M.; Challis, G. L. Nat. Chem. Biol. 2005, 1, 265–269. doi:10.1038/nchembio731 |
| 21. | Barona-Gómez, F.; Wong, U.; Giannakopulos, A. E.; Derrick, P. J.; Challis, G. L. J. Am. Chem. Soc. 2004, 126, 16282–16283. doi:10.1021/ja045774k |
| 22. | Thariath, A.; Socha, D.; Valvano, M. A.; Viswanatha, T. J. Bacteriol. 1993, 175, 589–596. doi:10.1128/jb.175.3.589-596.1993 |
| 12. | Oluwabusola, E. T.; Adebisi, O. O.; Reyes, F.; Acquah, K. S.; de la Cruz, M.; Mweetwa, L. L.; Rajakulendran, J. E.; Warner, D. F.; Hai, D.; Ebel, R.; Jaspars, M. Beilstein J. Org. Chem. 2021, 17, 2390–2398. doi:10.3762/bjoc.17.156 |
| 17. | Heemstra, J. R., Jr.; Walsh, C. T.; Sattely, E. S. J. Am. Chem. Soc. 2009, 131, 15317–15329. doi:10.1021/ja9056008 |
| 13. | Igarashi, Y.; Ge, Y.; Zhou, T.; Sharma, A. R.; Harunari, E.; Oku, N.; Trianto, A. Beilstein J. Org. Chem. 2022, 18, 110–119. doi:10.3762/bjoc.18.12 |
| 15. | Kreutzer, M. F.; Kage, H.; Nett, M. J. Am. Chem. Soc. 2012, 134, 5415–5422. doi:10.1021/ja300620z |
| 16. | Kreutzer, M. F.; Nett, M. Org. Biomol. Chem. 2012, 10, 9338–9343. doi:10.1039/c2ob26296g |
© 2022 Steinmetz et al.; licensee Beilstein-Institut.
This is an open access article licensed under the terms of the Beilstein-Institut Open Access License Agreement (https://www.beilstein-journals.org/bjoc/terms), which is identical to the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0). The reuse of material under this license requires that the author(s), source and license are credited. Third-party material in this article could be subject to other licenses (typically indicated in the credit line), and in this case, users are required to obtain permission from the license holder to reuse the material.