Abstract
Lipid II is an essential glycolipid found in bacteria. Accessing this valuable cell wall precursor is important both for studying cell wall synthesis and for studying/identifying novel antimicrobial compounds. Herein, we describe optimizations to the modular chemical synthesis of lipid II and unnatural analogues. In particular, the glycosylation step, a critical step in the formation of the central disaccharide unit (GlcNAc-MurNAc), was optimized. This was achieved by employing the use of glycosyl donors with diverse leaving groups. The key advantage of this approach lies in its adaptability, allowing for the generation of a wide array of analogues through the incorporation of alternative building blocks at different stages of synthesis.
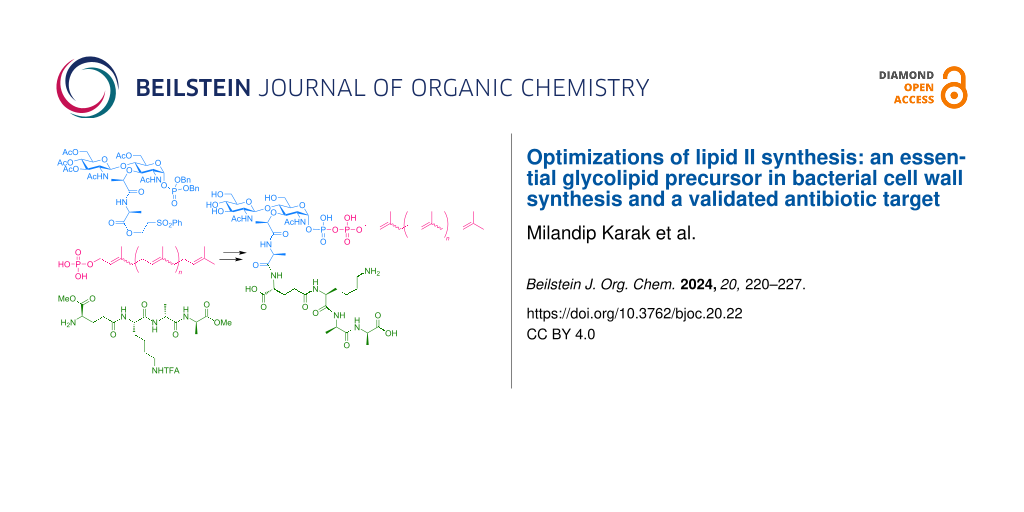
Graphical Abstract
Introduction
Lipid II (Figure 1) is an essential bacterial glycolipid involved in peptidoglycan biosynthesis [1]. It is synthesized on the inner leaflet of the cytoplasmic membrane, before translocation to the outer leaflet, where it is then used as the monomeric building block of peptidoglycan biosynthesis. Lipid II is a validated antibiotic target for clinically prescribed antibiotics including vancomycin and ramoplanin [2]. It is also the target for a host of other antimicrobials (mostly non-ribosomal peptides), including the tridecaptins [3], nisin [4], teixobactin [5], clovibactin [6], malacidin [7], and cilagicin [8].
Figure 1: Structure of lipid II, with variable positions shown in red and antimicrobial-binding motifs highlighted with blue arcs. R1 = H or Ac; R2 = H or Ac; R3 = OH, OMe or NH2; R4 = H or COOH; R5 = Gly5, Ala2, Ala-Ser/Ala or ᴅ-Asp; R6 = OH, OMe or NH2. These structural modifications are described in detail by Münch and co-workers [9]. For more details on lipid II-binding antimicrobials, see recent review by Buijs and co-workers [2].
Figure 1: Structure of lipid II, with variable positions shown in red and antimicrobial-binding motifs highli...
Despite significant progress in the chemical synthesis of lipid II and its analogues, the scarcity of these compounds and their limited structural diversity present significant obstacles to in-depth explorations of their intricate structural and functional characteristics. This scarcity issue is further exacerbated by an overwhelming demand that far exceeds existing supply capacities. To date, the chemical, chemoenzymatic, or biochemical synthesis of lipid II and its variants has been achieved by several research groups [10-27]. Nonetheless, considering the current state of knowledge, the chemical synthesis approach emerges as a more viable strategy in contrast to other methodologies, as it offers the potential to generate ample quantities of lipid II analogues suitable for high-throughput screening endeavors. In recent years, a major focus of the Cochrane lab has been the chemical synthesis of bacterial polyprenyls to study the mechanism of action of antimicrobial peptides that kill bacteria through binding to these polyprenyls [21,28-34]. Lipid II has been of particular interest, and during our synthesis of multiple different lipid II analogues, we have developed several optimizations, which we describe herein. The base lipid II syntheses upon which optimizations were made are our previously reported syntheses of Gram-negative lipid II in 2016 [20] and Gram-positive lipid II (11) in 2018 [23]. Building upon these synthetic strategies we have achieved noteworthy enhancements in glycosylation conditions, including improvements in reaction time and yields. This approach enables the systematic assembly of lipid II and analogues that contain shorter polyprenyl chains, specifically farnesyl (C15), geranylgeranyl (C20), and solanesyl (C45). Such short chain analogues are valuable in several applications due to their improved solubility in aqueous systems. Assembly is achieved by integrating distinct carbohydrate, peptide, and polyprenyl phosphate building blocks. This modular synthetic method allows for the strategic substitution of constituent building blocks at different synthetic stages and provides a practical avenue for producing substantial amounts of lipid II analogues. Consequently, this approach offers a more feasible means of addressing the demands associated with biophysical screening pursuits.
Prior research in the field of total synthesis of lipid II has elucidated that specific combinations of protecting groups on glycosyl acceptors and donors, as represented by compounds 1a and 2a in Figure 2, are proficient in the efficient generation of lipid II disaccharide [35,36]. Subsequently, significant endeavors have been directed towards the exploration of glycosyl donors, such as N-phthaloyl 3,4,6-O-triacetyl-2-deoxy-2-amino-ᴅ-glucopyranosyl-1-bromide, N-2,2,2-trichloroethoxycarbonyl-3,4,6-O-triacetyl-2-deoxy-2-amino-ᴅ-glucopyranosyl-1-bromide, and N-phthaloyl-2-deoxy-2-amino-3,4,6-O-triacetate-ᴅ-glucopyranosyl-1-(2,2,2-trichloroacetoimidate), all of which have proven successful in disaccharide synthesis alongside C6-protected acceptors (2a or 2b in Figure 2) [10,11,14,15,37,38]. More recently, an innovative one-pot glycosylation approach using a (2,6-dichloro-4-methoxyphenyl)(2,4-dichlorophenyl)-protected glycosyl acceptor has been developed, demonstrating satisfactory stability under Schmidt glycosylation conditions [18]. In general, the outcome of glycosylation hinges on the specific pairing of glycosyl donors and glycosyl acceptors employed in the reaction. Notably, when glycosyl donors such as 1e–g, featuring acyl group protection at the C2 position, are combined with acceptors like 2b, which have acyl groups protecting the C6 position, the reaction kinetics become sluggish, resulting in low conversion rates or no conversion [36,39].
Figure 2: List of i) glycosyl donors and ii) glycosyl acceptors used in this study.
Figure 2: List of i) glycosyl donors and ii) glycosyl acceptors used in this study.
Results and Discussion
In our studies, the initial glycosyl donors and acceptors (Figure 2; compounds 1a–g and 2a,b) were synthesized using established procedures from the literature, commencing with ᴅ-glucosamine and benzyl 2-acetamido-4,6-O-benzylidene-2-deoxy-α-ᴅ-glucopyranoside as the starting materials, respectively [40-43]. Imidate donors 1a and 1e were obtained exclusively as α-anomers, and 1b and 1g as a 1:1 α:β mixture which were then purified to give the desired α-anomers. Thioglycosides 1c, 1d, and 1f were isolated purely as β-anomers due to anchimeric assistance from the C2 N-acetyl or N-Troc groups. In glycosyl acceptors, the first amino acid of the lipid II pentapeptide, Ala, was incorporated as a 2-(phenylsulfonyl)ethyl ester, as previously reported by Saha and co-workers [44]. This modification prevents a deleterious side reaction occurring, wherein during glycosylation, muramic acid esters undergo a 6-exo-trig cyclization with the 4-OH group. Comprehensive experimental protocols detailing the preparation of these glycosyl donors can be found in Supporting Information File 1.
Next, we conducted an extended investigation into glycosylation, employing a diverse range of glycosyl donors (1a–g) and acceptors (2a and 2b), and the comprehensive results are presented in Table 1. Initially, our approach was guided by the established protocols of Kurosu et al., which had previously demonstrated effectiveness in glycosylating glycosyl trichloroacetimidate 1a and C6-benzylated MurNAc derivative 2a [18]. Despite our efforts to optimize the yield of the target product 3a, involving modifications to reaction conditions such as transitioning from 0 °C to room temperature and extending the reaction duration from 3 to 24 hours, we did not observe the anticipated enhancements (51% yield, entry 1, Table 1). This trend persisted when we attempted glycosylation between C6-acetylated MurNAc derivative 2b and 1a, where the desired product 3b remained elusive (Table 1, entry 2). In fact, glycosyl acceptor 2b failed to yield the desired glycosylation product 3d under the conditions tested (Table 1, entries 7 and 8). Moderate yields of 3a were achieved when using glycosyl donors such as 1b–d under standard conditions A or B (Table 1, entries 3–5). Notably, both Troc-protected thio-donors 1c,d exhibited similar behavior in terms of yield. Unfortunately, no target product 3c was obtained under standard glycosylation conditions A or B when C2-acetamido glycosyl donors (e.g., 1e–g) were subjected to the glycosylation reaction (Table 1, entries 6, 8, and 9). A slight improvement in the yield of 3a was observed when switching from TMSOTf to TfOH as the activator (Table 1, entry 5 vs entry 10). However, substituting TMSOTf with BF3·OEt2 did not yield any target product 3a (Table 1, entry 3 vs entry 12). In our observations, we initially noted that at room temperature, the degradation rate of glycosyl donor 1a exceeded the rate of product formation. This led to a complex mixture consisting of the target product 3a, acceptor 2a, and various degraded products of donor 1a. This situation posed challenges, as even prolonged reaction times did not enhance the product yield, and the subsequent purification of the target product became a difficult task. However, when we conducted the reaction at lower temperatures, the degradation of glycosyl donor 1a slowed down, and the reaction proceeded at a moderate rate. Eventually, we found that the utilization of extra equivalents of 1a and activators, following conditions akin to those employed by Kurosu, resulted in a significant boost in the yield of the target product to 68% (Table 1, entry 11).
Table 1: Optimization of the glycosylation conditions.a
|
|
|||||
| Entry | Donor | Acceptor | Deviation from std. conditions | Product | Yield (%) |
| 1 | 1a | 2a | conditions A | 3a | 51 |
| 2 | 1a | 2b | conditions A | 3b | 0 |
| 3 | 1b | 2a | conditions A | 3a | 29 |
| 4 | 1c | 2a | conditions B | 3a | 46 |
| 5 | 1d | 2a | conditions B | 3a | 43 |
| 6 | 1e | 2a | conditions A | 3c | 0 |
| 7 | 1e | 2b | conditions A | 3d | 0 |
| 8 | 1f | 2b | conditions B | 3d | 0 |
| 9 | 1g | 2a | conditions A | 3c | 0 |
| 10 | 1d | 2a | TfOH, NIS, 4 Å MS, CH2Cl2, −40 to 0 °C, 4 h | 3a | 50 |
| 11 | 1a | 2a | TMSOTf, 4 Å MS, CH2Cl2, 0 °C, 3 h; then, added 2 equiv 1a, 1 equiv TMSOTf, 0 °C, 4 h | 3a | 68 |
| 12 | 1b | 2a | BF3·OEt2, 4 Å MS, CH2Cl2, 0 °C to rt, 24 h | 3a | 0 |
aTMSOTf: trimethylsilyl trifluoromethanesulfonate, MS: molecular sieves, NIS: N-iodosuccinimide, Ac: acetyl, Bn: benzyl, Troc: 2,2,2-trichloroethoxycarbonyl.
Next, a comprehensive synthetic strategy for the preparation of α-phosphoryl GlcNAc-MurNAc-pentapeptide 7, based on established protocols with minor adjustments was completed (Scheme 1) [10,11]. After the successful glycosylation reaction, disaccharide 3a, protected with C2-Troc and C6-benzyl groups, was efficiently deprotected under acidic conditions using ZnCl2/Zn, followed by in situ re-acetylation of the C2-amino group and C6-alcohol with acetic anhydride, resulting in the formation of disaccharide 4 in a one-pot fashion. The anomeric benzyl protecting group in disaccharide 4 was then removed via a Pd/C-catalyzed hydrogenation reaction, producing a mixture of α/β-anomers of compound 5. It is noteworthy to mention that the benzyl ether in compound 4 exhibited successful cleavage upon treatment with sodium bromate/sodium dithionite in ethyl acetate/water, while other protecting functionalities like acetyl and phenylsulfonylethyl ester groups remained intact [45]. The ratio of α/β-anomers in compound 5 was found to be influenced by the reaction conditions, consistently favoring the β-anomer. Further transformation of compound 5 involved α-selective phosphite formation using dibenzyl N,N-diisopropylphosphoramidite and 5-(ethylthio)-1H-tetrazole. The resulting α-phosphite intermediate was then oxidized with hydrogen peroxide to yield dibenzyl α-phosphate 6, achieving an overall yield of 89% for these two steps. Removal of the 2-(phenylsulfonyl)ethanol protecting group in compound 6 was successfully achieved through treatment with 1,8-diazabicyclo[5.4.0]undec-7-ene, leading to the formation of the α-phosphoryl GlcNAc-MurNAc-monopeptide derivative. Subsequently, coupling this intermediate with tetrapeptide, TFA·H-ʟ-Ala-γ-ᴅ-Glu(OMe)-ʟ-Lys(COCF3)-ᴅ-Ala-ᴅ-Ala-OMe under mild conditions resulted in the synthesis of dibenzyl α-phosphoryl GlcNAc-MurNAc-pentapeptide 7 (see Supporting Information File 1 for comprehensive information on the synthesis details of the tetrapeptide). To avoid loss of valuable material through HPLC purification, crude 7 is used directly in the next step, and purification performed after the final prenyl phosphate coupling and global deprotection.
Scheme 1: Synthesis of disaccharide pentapeptide core 7.
Scheme 1: Synthesis of disaccharide pentapeptide core 7.
Finally, the benzyl-protecting groups in compound 7 were cleaved via hydrogenolysis, followed by co-evaporation of the resulting crude product in pyridine. This yielded a monopyridyl salt, setting the stage for the final lipid coupling and deprotection sequence. To establish the vital lipid diphosphate linkage, we employed the phosphoroimidazolidate method, as previously utilized in other lipid II total syntheses [10,11]. The monopyridyl α-phosphoryl GlcNAc-MurNAc-pentapeptide was activated with CDI, with excess CDI being neutralized using anhydrous methanol. The resulting phosphoroimidazolidate mixture underwent a cross-coupling reaction with prenyl monophosphates [46] in DMF/THF over a four-day period, yielding fully protected versions of lipid II and its analogues. Subsequent global deprotection reactions, using aqueous NaOH, led to the formation of lipid II (11), with an overall yield of 16% (from compound 7) following reversed-phase HPLC purification (Scheme 2). Similarly, farnesyl, geranylgeranyl, and solanesyl-lipid II analogues 8–10 were synthesized with overall yields of 13%, 21%, and 11%, respectively, using the corresponding prenyl phosphates (Scheme 2).
Scheme 2: Synthesis of lipid II (11) and its analogues 8–10.
Scheme 2: Synthesis of lipid II (11) and its analogues 8–10.
Conclusion
In conclusion, we have successfully optimized a modular approach for the synthesis of lipid II and its analogues, including variants with distinct prenyl-chain lengths. The key to this methodology lies in the optimization of glycosylation conditions, utilizing readily available glycosyl donors, which is a pivotal step in constructing the central disaccharide unit. The adaptability of our method is showcased through the generation of new lipid II analogues, such as geranylgeranyl and solanesyl lipid II analogues, which involve the incorporation of distinct prenyl monophosphates during the final phases of the synthesis. Thus, this strategy holds considerable promise for advancing the synthesis of a diverse range of lipid II analogues, opening avenues for further exploration into their biophysical characteristics, as well as their interactions with antibiotics.
Supporting Information
| Supporting Information File 1: Experimental procedures, characterization data, and selected copies of 1H, 13C, and 31P NMR spectra. | ||
| Format: PDF | Size: 3.7 MB | Download |
Acknowledgements
We extend our gratitude to Professor Alethea Tabor and Professor Stefan Howorka from University College London for their valuable collaboration on this project. We also express our appreciation to the dedicated technical team at CCE-QUB for their unwavering technical support throughout this project.
Data Availability Statement
The data that supports the findings of this study is available from the corresponding author upon reasonable request.
References
-
Egan, A. J. F.; Errington, J.; Vollmer, W. Nat. Rev. Microbiol. 2020, 18, 446–460. doi:10.1038/s41579-020-0366-3
Return to citation in text: [1] -
Buijs, N. P.; Matheson, E. J.; Cochrane, S. A.; Martin, N. I. Chem. Commun. 2023, 59, 7685–7703. doi:10.1039/d3cc01070h
Return to citation in text: [1] [2] -
Bann, S. J.; Ballantine, R. D.; Cochrane, S. A. RSC Med. Chem. 2021, 12, 538–551. doi:10.1039/d0md00413h
Return to citation in text: [1] -
Medeiros-Silva, J.; Jekhmane, S.; Paioni, A. L.; Gawarecka, K.; Baldus, M.; Swiezewska, E.; Breukink, E.; Weingarth, M. Nat. Commun. 2018, 9, 3963. doi:10.1038/s41467-018-06314-x
Return to citation in text: [1] -
Medeiros‐Silva, J.; Jekhmane, S.; Breukink, E.; Weingarth, M. ChemBioChem 2019, 20, 1731–1738. doi:10.1002/cbic.201800796
Return to citation in text: [1] -
Shukla, R.; Peoples, A. J.; Ludwig, K. C.; Maity, S.; Derks, M. G. N.; De Benedetti, S.; Krueger, A. M.; Vermeulen, B. J. A.; Harbig, T.; Lavore, F.; Kumar, R.; Honorato, R. V.; Grein, F.; Nieselt, K.; Liu, Y.; Bonvin, A. M. J. J.; Baldus, M.; Kubitscheck, U.; Breukink, E.; Achorn, C.; Nitti, A.; Schwalen, C. J.; Spoering, A. L.; Ling, L. L.; Hughes, D.; Lelli, M.; Roos, W. H.; Lewis, K.; Schneider, T.; Weingarth, M. Cell 2023, 186, 4059–4073. doi:10.1016/j.cell.2023.07.038
Return to citation in text: [1] -
Hover, B. M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J. G.; Ternei, M. A.; Maniko, J.; Estrela, A. B.; Molina, H.; Park, S.; Perlin, D. S.; Brady, S. F. Nat. Microbiol. 2018, 3, 415–422. doi:10.1038/s41564-018-0110-1
Return to citation in text: [1] -
Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Brady, S. F. Science 2022, 376, 991–996. doi:10.1126/science.abn4213
Return to citation in text: [1] -
Münch, D.; Sahl, H.-G. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 3062–3071. doi:10.1016/j.bbamem.2015.04.014
Return to citation in text: [1] -
Schwartz, B.; Markwalder, J. A.; Wang, Y. J. Am. Chem. Soc. 2001, 123, 11638–11643. doi:10.1021/ja0166848
Return to citation in text: [1] [2] [3] [4] -
VanNieuwenhze, M. S.; Mauldin, S. C.; Zia-Ebrahimi, M.; Winger, B. E.; Hornback, W. J.; Saha, S. L.; Aikins, J. A.; Blaszczak, L. C. J. Am. Chem. Soc. 2002, 124, 3656–3660. doi:10.1021/ja017386d
Return to citation in text: [1] [2] [3] [4] -
Liu, H.; Wong, C.-H. Bioorg. Med. Chem. 2006, 14, 7187–7195. doi:10.1016/j.bmc.2006.06.058
Return to citation in text: [1] -
Liu, C.-Y.; Guo, C.-W.; Chang, Y.-F.; Wang, J.-T.; Shih, H.-W.; Hsu, Y.-F.; Chen, C.-W.; Chen, S.-K.; Wang, Y.-C.; Cheng, T.-J. R.; Ma, C.; Wong, C.-H.; Fang, J.-M.; Cheng, W.-C. Org. Lett. 2010, 12, 1608–1611. doi:10.1021/ol100338v
Return to citation in text: [1] -
Shih, H.-W.; Chen, K.-T.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 4600–4603. doi:10.1021/ol201806d
Return to citation in text: [1] [2] -
Meng, F.-C.; Chen, K.-T.; Huang, L.-Y.; Shih, H.-W.; Chang, H.-H.; Nien, F.-Y.; Liang, P.-H.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 5306–5309. doi:10.1021/ol2021687
Return to citation in text: [1] [2] -
Chen, K.-T.; Kuan, Y.-C.; Fu, W.-C.; Liang, P.-H.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Chem. – Eur. J. 2013, 19, 834–838. doi:10.1002/chem.201203251
Return to citation in text: [1] -
Huang, L.-Y.; Huang, S.-H.; Chang, Y.-C.; Cheng, W.-C.; Cheng, T.-J. R.; Wong, C.-H. Angew. Chem., Int. Ed. 2014, 53, 8060–8065. doi:10.1002/anie.201402313
Return to citation in text: [1] -
Mitachi, K.; Mohan, P.; Siricilla, S.; Kurosu, M. Chem. – Eur. J. 2014, 20, 4554–4558. doi:10.1002/chem.201400307
Return to citation in text: [1] [2] [3] -
Lin, C.-K.; Chen, K.-T.; Hu, C.-M.; Yun, W.-Y.; Cheng, W.-C. Chem. – Eur. J. 2015, 21, 7511–7519. doi:10.1002/chem.201406629
Return to citation in text: [1] -
Cochrane, S. A.; Findlay, B.; Bakhtiary, A.; Acedo, J. Z.; Rodriguez-Lopez, E. M.; Mercier, P.; Vederas, J. C. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11561–11566. doi:10.1073/pnas.1608623113
Return to citation in text: [1] [2] -
Bakhtiary, A.; Cochrane, S. A.; Mercier, P.; McKay, R. T.; Miskolzie, M.; Sit, C. S.; Vederas, J. C. J. Am. Chem. Soc. 2017, 139, 17803–17810. doi:10.1021/jacs.7b04728
Return to citation in text: [1] [2] -
Katsuyama, A.; Sato, K.; Yakushiji, F.; Matsumaru, T.; Ichikawa, S. Chem. Pharm. Bull. 2018, 66, 84–95. doi:10.1248/cpb.c17-00828
Return to citation in text: [1] -
Dong, Y. Y.; Wang, H.; Pike, A. C. W.; Cochrane, S. A.; Hamedzadeh, S.; Wyszyński, F. J.; Bushell, S. R.; Royer, S. F.; Widdick, D. A.; Sajid, A.; Boshoff, H. I.; Park, Y.; Lucas, R.; Liu, W.-M.; Lee, S. S.; Machida, T.; Minall, L.; Mehmood, S.; Belaya, K.; Liu, W.-W.; Chu, A.; Shrestha, L.; Mukhopadhyay, S. M. M.; Strain-Damerell, C.; Chalk, R.; Burgess-Brown, N. A.; Bibb, M. J.; Barry, C. E., III; Robinson, C. V.; Beeson, D.; Davis, B. G.; Carpenter, E. P. Cell 2018, 175, 1045–1058. doi:10.1016/j.cell.2018.10.037
Return to citation in text: [1] [2] -
Wingen, L. M.; Rausch, M.; Schneider, T.; Menche, D. J. Org. Chem. 2020, 85, 10206–10215. doi:10.1021/acs.joc.0c01004
Return to citation in text: [1] -
Cochrane, S. A.; Lohans, C. T. Eur. J. Med. Chem. 2020, 194, 112262. doi:10.1016/j.ejmech.2020.112262
Return to citation in text: [1] -
Hsu, T.-W.; Fang, J.-M. Analyst (Cambridge, U. K.) 2021, 146, 5843–5847. doi:10.1039/d1an00941a
Return to citation in text: [1] -
Katsuyama, A.; Yakushiji, F.; Ichikawa, S. Tetrahedron Lett. 2021, 73, 153101. doi:10.1016/j.tetlet.2021.153101
Return to citation in text: [1] -
Kotsogianni, I.; Wood, T. M.; Alexander, F. M.; Cochrane, S. A.; Martin, N. I. ACS Infect. Dis. 2021, 7, 2612–2619. doi:10.1021/acsinfecdis.1c00316
Return to citation in text: [1] -
Baker, B. R.; Ives, C. M.; Bray, A.; Caffrey, M.; Cochrane, S. A. Eur. J. Med. Chem. 2021, 210, 113062. doi:10.1016/j.ejmech.2020.113062
Return to citation in text: [1] -
Cochrane, R. V. K.; Alexander, F. M.; Boland, C.; Fetics, S. K.; Caffrey, M.; Cochrane, S. A. Chem. Commun. 2020, 56, 8603–8606. doi:10.1039/d0cc03388j
Return to citation in text: [1] -
Chiorean, S.; Antwi, I.; Carney, D. W.; Kotsogianni, I.; Giltrap, A. M.; Alexander, F. M.; Cochrane, S. A.; Payne, R. J.; Martin, N. I.; Henninot, A.; Vederas, J. C. ChemBioChem 2020, 21, 789–792. doi:10.1002/cbic.201900504
Return to citation in text: [1] -
Bann, S. J.; Ballantine, R. D.; McCallion, C. E.; Qian, P.-Y.; Li, Y.-X.; Cochrane, S. A. J. Med. Chem. 2019, 62, 10466–10472. doi:10.1021/acs.jmedchem.9b01078
Return to citation in text: [1] -
Ballantine, R. D.; McCallion, C. E.; Nassour, E.; Tokajian, S.; Cochrane, S. A. Med. Chem. Commun. 2019, 10, 484–487. doi:10.1039/c9md00031c
Return to citation in text: [1] -
Ballantine, R. D.; Li, Y.-X.; Qian, P.-Y.; Cochrane, S. A. Chem. Commun. 2018, 54, 10634–10637. doi:10.1039/c8cc05790g
Return to citation in text: [1] -
Termin, A.; Schmidt, R. R. Liebigs Ann. Chem. 1992, 527–533. doi:10.1002/jlac.199219920191
Return to citation in text: [1] -
Zhang, Z.; Ollmann, I. R.; Ye, X.-S.; Wischnat, R.; Baasov, T.; Wong, C.-H. J. Am. Chem. Soc. 1999, 121, 734–753. doi:10.1021/ja982232s
Return to citation in text: [1] [2] -
Hitchcock, S. A.; Eid, C. N.; Aikins, J. A.; Zia-Ebrahimi, M.; Blaszczak, L. C. J. Am. Chem. Soc. 1998, 120, 1916–1917. doi:10.1021/ja973172d
Return to citation in text: [1] -
Zhang, Y.; Fechter, E. J.; Wang, T.-S. A.; Barrett, D.; Walker, S.; Kahne, D. E. J. Am. Chem. Soc. 2007, 129, 3080–3081. doi:10.1021/ja069060g
Return to citation in text: [1] -
Ritter, T. K.; Mong, K.-K. T.; Liu, H.; Nakatani, T.; Wong, C.-H. Angew. Chem., Int. Ed. 2003, 42, 4657–4660. doi:10.1002/anie.200351534
Return to citation in text: [1] -
Lioux, T.; Busson, R.; Rozenski, J.; Nguyen-Distèche, M.; Frère, J.-M.; Herdewijn, P. Collect. Czech. Chem. Commun. 2005, 70, 1615–1641. doi:10.1135/cccc20051615
Return to citation in text: [1] -
Grann Hansen, S.; Skrydstrup, T. Eur. J. Org. Chem. 2007, 3392–3401. doi:10.1002/ejoc.200700048
Return to citation in text: [1] -
Subramanian, V.; Moumé-Pymbock, M.; Hu, T.; Crich, D. J. Org. Chem. 2011, 76, 3691–3709. doi:10.1021/jo102411j
Return to citation in text: [1] -
Xu, C.; Liu, H.; Li, X. Carbohydr. Res. 2011, 346, 1149–1153. doi:10.1016/j.carres.2011.03.033
Return to citation in text: [1] -
Saha, S. L.; Van Nieuwenhze, M. S.; Hornback, W. J.; Aikins, J. A.; Blaszczak, L. C. Org. Lett. 2001, 3, 3575–3577. doi:10.1021/ol016692t
Return to citation in text: [1] -
Adinolfi, M.; Barone, G.; Guariniello, L.; Iadonisi, A. Tetrahedron Lett. 1999, 40, 8439–8441. doi:10.1016/s0040-4039(99)01756-6
Return to citation in text: [1] -
Lira, L. M.; Vasilev, D.; Pilli, R. A.; Wessjohann, L. A. Tetrahedron Lett. 2013, 54, 1690–1692. doi:10.1016/j.tetlet.2013.01.059
Return to citation in text: [1]
| 1. | Egan, A. J. F.; Errington, J.; Vollmer, W. Nat. Rev. Microbiol. 2020, 18, 446–460. doi:10.1038/s41579-020-0366-3 |
| 5. | Medeiros‐Silva, J.; Jekhmane, S.; Breukink, E.; Weingarth, M. ChemBioChem 2019, 20, 1731–1738. doi:10.1002/cbic.201800796 |
| 35. | Termin, A.; Schmidt, R. R. Liebigs Ann. Chem. 1992, 527–533. doi:10.1002/jlac.199219920191 |
| 36. | Zhang, Z.; Ollmann, I. R.; Ye, X.-S.; Wischnat, R.; Baasov, T.; Wong, C.-H. J. Am. Chem. Soc. 1999, 121, 734–753. doi:10.1021/ja982232s |
| 4. | Medeiros-Silva, J.; Jekhmane, S.; Paioni, A. L.; Gawarecka, K.; Baldus, M.; Swiezewska, E.; Breukink, E.; Weingarth, M. Nat. Commun. 2018, 9, 3963. doi:10.1038/s41467-018-06314-x |
| 10. | Schwartz, B.; Markwalder, J. A.; Wang, Y. J. Am. Chem. Soc. 2001, 123, 11638–11643. doi:10.1021/ja0166848 |
| 11. | VanNieuwenhze, M. S.; Mauldin, S. C.; Zia-Ebrahimi, M.; Winger, B. E.; Hornback, W. J.; Saha, S. L.; Aikins, J. A.; Blaszczak, L. C. J. Am. Chem. Soc. 2002, 124, 3656–3660. doi:10.1021/ja017386d |
| 14. | Shih, H.-W.; Chen, K.-T.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 4600–4603. doi:10.1021/ol201806d |
| 15. | Meng, F.-C.; Chen, K.-T.; Huang, L.-Y.; Shih, H.-W.; Chang, H.-H.; Nien, F.-Y.; Liang, P.-H.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 5306–5309. doi:10.1021/ol2021687 |
| 37. | Hitchcock, S. A.; Eid, C. N.; Aikins, J. A.; Zia-Ebrahimi, M.; Blaszczak, L. C. J. Am. Chem. Soc. 1998, 120, 1916–1917. doi:10.1021/ja973172d |
| 38. | Zhang, Y.; Fechter, E. J.; Wang, T.-S. A.; Barrett, D.; Walker, S.; Kahne, D. E. J. Am. Chem. Soc. 2007, 129, 3080–3081. doi:10.1021/ja069060g |
| 3. | Bann, S. J.; Ballantine, R. D.; Cochrane, S. A. RSC Med. Chem. 2021, 12, 538–551. doi:10.1039/d0md00413h |
| 20. | Cochrane, S. A.; Findlay, B.; Bakhtiary, A.; Acedo, J. Z.; Rodriguez-Lopez, E. M.; Mercier, P.; Vederas, J. C. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11561–11566. doi:10.1073/pnas.1608623113 |
| 2. | Buijs, N. P.; Matheson, E. J.; Cochrane, S. A.; Martin, N. I. Chem. Commun. 2023, 59, 7685–7703. doi:10.1039/d3cc01070h |
| 23. | Dong, Y. Y.; Wang, H.; Pike, A. C. W.; Cochrane, S. A.; Hamedzadeh, S.; Wyszyński, F. J.; Bushell, S. R.; Royer, S. F.; Widdick, D. A.; Sajid, A.; Boshoff, H. I.; Park, Y.; Lucas, R.; Liu, W.-M.; Lee, S. S.; Machida, T.; Minall, L.; Mehmood, S.; Belaya, K.; Liu, W.-W.; Chu, A.; Shrestha, L.; Mukhopadhyay, S. M. M.; Strain-Damerell, C.; Chalk, R.; Burgess-Brown, N. A.; Bibb, M. J.; Barry, C. E., III; Robinson, C. V.; Beeson, D.; Davis, B. G.; Carpenter, E. P. Cell 2018, 175, 1045–1058. doi:10.1016/j.cell.2018.10.037 |
| 9. | Münch, D.; Sahl, H.-G. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 3062–3071. doi:10.1016/j.bbamem.2015.04.014 |
| 10. | Schwartz, B.; Markwalder, J. A.; Wang, Y. J. Am. Chem. Soc. 2001, 123, 11638–11643. doi:10.1021/ja0166848 |
| 11. | VanNieuwenhze, M. S.; Mauldin, S. C.; Zia-Ebrahimi, M.; Winger, B. E.; Hornback, W. J.; Saha, S. L.; Aikins, J. A.; Blaszczak, L. C. J. Am. Chem. Soc. 2002, 124, 3656–3660. doi:10.1021/ja017386d |
| 12. | Liu, H.; Wong, C.-H. Bioorg. Med. Chem. 2006, 14, 7187–7195. doi:10.1016/j.bmc.2006.06.058 |
| 13. | Liu, C.-Y.; Guo, C.-W.; Chang, Y.-F.; Wang, J.-T.; Shih, H.-W.; Hsu, Y.-F.; Chen, C.-W.; Chen, S.-K.; Wang, Y.-C.; Cheng, T.-J. R.; Ma, C.; Wong, C.-H.; Fang, J.-M.; Cheng, W.-C. Org. Lett. 2010, 12, 1608–1611. doi:10.1021/ol100338v |
| 14. | Shih, H.-W.; Chen, K.-T.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 4600–4603. doi:10.1021/ol201806d |
| 15. | Meng, F.-C.; Chen, K.-T.; Huang, L.-Y.; Shih, H.-W.; Chang, H.-H.; Nien, F.-Y.; Liang, P.-H.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Org. Lett. 2011, 13, 5306–5309. doi:10.1021/ol2021687 |
| 16. | Chen, K.-T.; Kuan, Y.-C.; Fu, W.-C.; Liang, P.-H.; Cheng, T.-J. R.; Wong, C.-H.; Cheng, W.-C. Chem. – Eur. J. 2013, 19, 834–838. doi:10.1002/chem.201203251 |
| 17. | Huang, L.-Y.; Huang, S.-H.; Chang, Y.-C.; Cheng, W.-C.; Cheng, T.-J. R.; Wong, C.-H. Angew. Chem., Int. Ed. 2014, 53, 8060–8065. doi:10.1002/anie.201402313 |
| 18. | Mitachi, K.; Mohan, P.; Siricilla, S.; Kurosu, M. Chem. – Eur. J. 2014, 20, 4554–4558. doi:10.1002/chem.201400307 |
| 19. | Lin, C.-K.; Chen, K.-T.; Hu, C.-M.; Yun, W.-Y.; Cheng, W.-C. Chem. – Eur. J. 2015, 21, 7511–7519. doi:10.1002/chem.201406629 |
| 20. | Cochrane, S. A.; Findlay, B.; Bakhtiary, A.; Acedo, J. Z.; Rodriguez-Lopez, E. M.; Mercier, P.; Vederas, J. C. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11561–11566. doi:10.1073/pnas.1608623113 |
| 21. | Bakhtiary, A.; Cochrane, S. A.; Mercier, P.; McKay, R. T.; Miskolzie, M.; Sit, C. S.; Vederas, J. C. J. Am. Chem. Soc. 2017, 139, 17803–17810. doi:10.1021/jacs.7b04728 |
| 22. | Katsuyama, A.; Sato, K.; Yakushiji, F.; Matsumaru, T.; Ichikawa, S. Chem. Pharm. Bull. 2018, 66, 84–95. doi:10.1248/cpb.c17-00828 |
| 23. | Dong, Y. Y.; Wang, H.; Pike, A. C. W.; Cochrane, S. A.; Hamedzadeh, S.; Wyszyński, F. J.; Bushell, S. R.; Royer, S. F.; Widdick, D. A.; Sajid, A.; Boshoff, H. I.; Park, Y.; Lucas, R.; Liu, W.-M.; Lee, S. S.; Machida, T.; Minall, L.; Mehmood, S.; Belaya, K.; Liu, W.-W.; Chu, A.; Shrestha, L.; Mukhopadhyay, S. M. M.; Strain-Damerell, C.; Chalk, R.; Burgess-Brown, N. A.; Bibb, M. J.; Barry, C. E., III; Robinson, C. V.; Beeson, D.; Davis, B. G.; Carpenter, E. P. Cell 2018, 175, 1045–1058. doi:10.1016/j.cell.2018.10.037 |
| 24. | Wingen, L. M.; Rausch, M.; Schneider, T.; Menche, D. J. Org. Chem. 2020, 85, 10206–10215. doi:10.1021/acs.joc.0c01004 |
| 25. | Cochrane, S. A.; Lohans, C. T. Eur. J. Med. Chem. 2020, 194, 112262. doi:10.1016/j.ejmech.2020.112262 |
| 26. | Hsu, T.-W.; Fang, J.-M. Analyst (Cambridge, U. K.) 2021, 146, 5843–5847. doi:10.1039/d1an00941a |
| 27. | Katsuyama, A.; Yakushiji, F.; Ichikawa, S. Tetrahedron Lett. 2021, 73, 153101. doi:10.1016/j.tetlet.2021.153101 |
| 8. | Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Brady, S. F. Science 2022, 376, 991–996. doi:10.1126/science.abn4213 |
| 21. | Bakhtiary, A.; Cochrane, S. A.; Mercier, P.; McKay, R. T.; Miskolzie, M.; Sit, C. S.; Vederas, J. C. J. Am. Chem. Soc. 2017, 139, 17803–17810. doi:10.1021/jacs.7b04728 |
| 28. | Kotsogianni, I.; Wood, T. M.; Alexander, F. M.; Cochrane, S. A.; Martin, N. I. ACS Infect. Dis. 2021, 7, 2612–2619. doi:10.1021/acsinfecdis.1c00316 |
| 29. | Baker, B. R.; Ives, C. M.; Bray, A.; Caffrey, M.; Cochrane, S. A. Eur. J. Med. Chem. 2021, 210, 113062. doi:10.1016/j.ejmech.2020.113062 |
| 30. | Cochrane, R. V. K.; Alexander, F. M.; Boland, C.; Fetics, S. K.; Caffrey, M.; Cochrane, S. A. Chem. Commun. 2020, 56, 8603–8606. doi:10.1039/d0cc03388j |
| 31. | Chiorean, S.; Antwi, I.; Carney, D. W.; Kotsogianni, I.; Giltrap, A. M.; Alexander, F. M.; Cochrane, S. A.; Payne, R. J.; Martin, N. I.; Henninot, A.; Vederas, J. C. ChemBioChem 2020, 21, 789–792. doi:10.1002/cbic.201900504 |
| 32. | Bann, S. J.; Ballantine, R. D.; McCallion, C. E.; Qian, P.-Y.; Li, Y.-X.; Cochrane, S. A. J. Med. Chem. 2019, 62, 10466–10472. doi:10.1021/acs.jmedchem.9b01078 |
| 33. | Ballantine, R. D.; McCallion, C. E.; Nassour, E.; Tokajian, S.; Cochrane, S. A. Med. Chem. Commun. 2019, 10, 484–487. doi:10.1039/c9md00031c |
| 34. | Ballantine, R. D.; Li, Y.-X.; Qian, P.-Y.; Cochrane, S. A. Chem. Commun. 2018, 54, 10634–10637. doi:10.1039/c8cc05790g |
| 7. | Hover, B. M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J. G.; Ternei, M. A.; Maniko, J.; Estrela, A. B.; Molina, H.; Park, S.; Perlin, D. S.; Brady, S. F. Nat. Microbiol. 2018, 3, 415–422. doi:10.1038/s41564-018-0110-1 |
| 6. | Shukla, R.; Peoples, A. J.; Ludwig, K. C.; Maity, S.; Derks, M. G. N.; De Benedetti, S.; Krueger, A. M.; Vermeulen, B. J. A.; Harbig, T.; Lavore, F.; Kumar, R.; Honorato, R. V.; Grein, F.; Nieselt, K.; Liu, Y.; Bonvin, A. M. J. J.; Baldus, M.; Kubitscheck, U.; Breukink, E.; Achorn, C.; Nitti, A.; Schwalen, C. J.; Spoering, A. L.; Ling, L. L.; Hughes, D.; Lelli, M.; Roos, W. H.; Lewis, K.; Schneider, T.; Weingarth, M. Cell 2023, 186, 4059–4073. doi:10.1016/j.cell.2023.07.038 |
| 2. | Buijs, N. P.; Matheson, E. J.; Cochrane, S. A.; Martin, N. I. Chem. Commun. 2023, 59, 7685–7703. doi:10.1039/d3cc01070h |
| 40. | Lioux, T.; Busson, R.; Rozenski, J.; Nguyen-Distèche, M.; Frère, J.-M.; Herdewijn, P. Collect. Czech. Chem. Commun. 2005, 70, 1615–1641. doi:10.1135/cccc20051615 |
| 41. | Grann Hansen, S.; Skrydstrup, T. Eur. J. Org. Chem. 2007, 3392–3401. doi:10.1002/ejoc.200700048 |
| 42. | Subramanian, V.; Moumé-Pymbock, M.; Hu, T.; Crich, D. J. Org. Chem. 2011, 76, 3691–3709. doi:10.1021/jo102411j |
| 43. | Xu, C.; Liu, H.; Li, X. Carbohydr. Res. 2011, 346, 1149–1153. doi:10.1016/j.carres.2011.03.033 |
| 18. | Mitachi, K.; Mohan, P.; Siricilla, S.; Kurosu, M. Chem. – Eur. J. 2014, 20, 4554–4558. doi:10.1002/chem.201400307 |
| 36. | Zhang, Z.; Ollmann, I. R.; Ye, X.-S.; Wischnat, R.; Baasov, T.; Wong, C.-H. J. Am. Chem. Soc. 1999, 121, 734–753. doi:10.1021/ja982232s |
| 39. | Ritter, T. K.; Mong, K.-K. T.; Liu, H.; Nakatani, T.; Wong, C.-H. Angew. Chem., Int. Ed. 2003, 42, 4657–4660. doi:10.1002/anie.200351534 |
| 10. | Schwartz, B.; Markwalder, J. A.; Wang, Y. J. Am. Chem. Soc. 2001, 123, 11638–11643. doi:10.1021/ja0166848 |
| 11. | VanNieuwenhze, M. S.; Mauldin, S. C.; Zia-Ebrahimi, M.; Winger, B. E.; Hornback, W. J.; Saha, S. L.; Aikins, J. A.; Blaszczak, L. C. J. Am. Chem. Soc. 2002, 124, 3656–3660. doi:10.1021/ja017386d |
| 46. | Lira, L. M.; Vasilev, D.; Pilli, R. A.; Wessjohann, L. A. Tetrahedron Lett. 2013, 54, 1690–1692. doi:10.1016/j.tetlet.2013.01.059 |
| 10. | Schwartz, B.; Markwalder, J. A.; Wang, Y. J. Am. Chem. Soc. 2001, 123, 11638–11643. doi:10.1021/ja0166848 |
| 11. | VanNieuwenhze, M. S.; Mauldin, S. C.; Zia-Ebrahimi, M.; Winger, B. E.; Hornback, W. J.; Saha, S. L.; Aikins, J. A.; Blaszczak, L. C. J. Am. Chem. Soc. 2002, 124, 3656–3660. doi:10.1021/ja017386d |
| 45. | Adinolfi, M.; Barone, G.; Guariniello, L.; Iadonisi, A. Tetrahedron Lett. 1999, 40, 8439–8441. doi:10.1016/s0040-4039(99)01756-6 |
| 44. | Saha, S. L.; Van Nieuwenhze, M. S.; Hornback, W. J.; Aikins, J. A.; Blaszczak, L. C. Org. Lett. 2001, 3, 3575–3577. doi:10.1021/ol016692t |
| 18. | Mitachi, K.; Mohan, P.; Siricilla, S.; Kurosu, M. Chem. – Eur. J. 2014, 20, 4554–4558. doi:10.1002/chem.201400307 |
© 2024 Karak et al.; licensee Beilstein-Institut.
This is an open access article licensed under the terms of the Beilstein-Institut Open Access License Agreement (https://www.beilstein-journals.org/bjoc/terms), which is identical to the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0). The reuse of material under this license requires that the author(s), source and license are credited. Third-party material in this article could be subject to other licenses (typically indicated in the credit line), and in this case, users are required to obtain permission from the license holder to reuse the material.