Abstract
Ten dipyridodiazepinone derivatives were synthesized and evaluated for their anti HIV-1 reverse transcriptase activity against wild-type and mutant type enzymes, K103N and Y181C. Two of them were found to be promising inhibitors for HIV-1 RT.
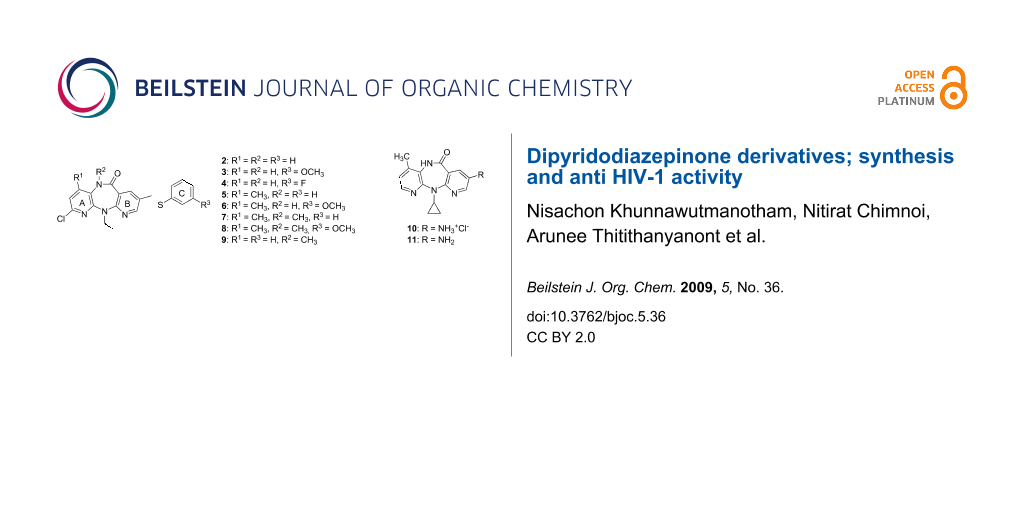
Graphical Abstract
Introduction
Dipyridodiazepinone nevirapine (1) [1] (Figure 1) is a potent non-nucleoside inhibitor of human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) and is approved as a therapeutic agent for the treatment of AIDS. In the clinic, nevirapine monotherapy results in relatively rapid drug resistance due to mutation of the RT enzyme. To develop a second-generation inhibitor with improved activity against the mutant RT enzyme, many efforts have been focused on the synthesis of dipyridodiazepinone derivatives [2-8]. On the basis of molecular modeling analysis on the wild-type (WT) and Y181C HIV-1 RT, it was found that the dipyridodiazepinone derivatives containing an unsubstituted lactam nitrogen and a 2-chloro-8-arylthiomethyl moiety, when compared with 9 [4] (Figure 2) as reference, are effective inhibitors of this mutant enzyme. Some dipyridodiazepinone derivatives containing an N-methylated of lactam also exhibited good potency against the WT enzyme. The 8-amino derivative of nevirapine and its hydrochloride salt also provided interesting potency. The first two compounds, 2 and 3, were synthesized and their virustatic and virucidal activities against HIV-1 subtype E were reported previously [9]. As part of our continuing efforts directed towards the development of potential HIV-1 RT inhibitors, we have extended the synthesis of dipyridodiazepinone derivatives (Figure 2) and evaluation of their activity against wild-type RT and K103N and Y181C mutant RT enzymes.
Figure 2: Structures of dipyridodiazepinone derivatives with promising anti-HIV activity.
Figure 2: Structures of dipyridodiazepinone derivatives with promising anti-HIV activity.
Results and Discussion
Synthesis of compounds 2–9
Compounds 2–9 were synthesized via efficient routes as shown in Scheme 1 and Scheme 2. The aminopyridinecarboxamide 15a (R = H) was prepared from 2-(ethylamino)-3-pyridinecarboxylic acid (13) and 3-amino-2,6-dichloropyridine (14) (Scheme 1) [9]. However, by using the same procedure to prepare aminopyridinecarboxamide 15b (R = CH3), only poor yields of 15b were obtained. Therefore, 15b (R = CH3) was prepared by formation of pyridinecarboxamide 17, obtained from 12 and 16 [10,11], followed by the displacement of 2′ chloro by the ethylamino group.
Scheme 1: Reagents and conditions: (a) EtNH2, 120 °C, 4 h, 99% (b) i) (COCl)2, benzene, DMF, rt, 1 h; ii) amine 14 or 16, dioxane, cyclohexane, pyridine, rt, 16 h, 80% (c) EtNH2, dioxane, 100 °C, 20 min, 47%.
Scheme 1: Reagents and conditions: (a) EtNH2, 120 °C, 4 h, 99% (b) i) (COCl)2, benzene, DMF, rt, 1 h; ii) ami...
Scheme 2: Reagents and conditions: (a) Br2, HOAc, KOAc, rt, 1 h; (b) NaHMDS, pyridine, 90 °C, 1 h; (c) CH2=CH–SnBu3, Pd(PPh3)4, DMF, 90 °C, 1 h; (d) O3, CH2Cl2/MeOH, −78 °C then PPh3, rt, 1 h; (e) NaBH4, THF, H2O, rt, 0.5 h; (f) SOCl2, CH2Cl2, Et3N, rt; (g) NaH, ArSH, DMF, rt, 1 h; (h) NaH, DMF, 50 °C, 0.5 h then MeI, rt, 0.5 h.
Scheme 2: Reagents and conditions: (a) Br2, HOAc, KOAc, rt, 1 h; (b) NaHMDS, pyridine, 90 °C, 1 h; (c) CH2=CH...
Afterwards, the aminopyridinecarboxamide intermediates 15 were treated as previously reported [4] to give 8-arylthiomethyldipyridodiazepinones, as shown in Scheme 2.
Aminopyridinecarboxamides 15 were regioselectively brominated to produce bromo compounds 18. The diazepinone ring was formed by treatment with sodium hexamethyldisilazane in pyridine to yield tricyclic compounds 19. Coupling of 19 with vinyltributyltin in the presence of tetrakis(triphenylphosphine) palladium(0) provided 8-vinyl compounds 20 which underwent ozonolysis to produce aldehydes 21 in good yields. The reduction of 21 with NaBH4 produced alcohols 22, which were converted to the corresponding chlorides 23 through treatment with thionyl chloride in dichloromethane. The reaction of 23a with thiophenolate, 3-methoxythiophenolate, and 3-fluorothiophenolate in N,N-dimethylformamide yielded 2, 3, and 4, respectively, whilst the reaction of 23b with thiophenolate and 3-methoxythiophenolate yielded 5 and 6, respectively. Methylation of the lactam nitrogen of 5 and 6 with methyl iodide provided 7 and 8. Compound 9 was also prepared via methylation of 2, which was used as the reference compound.
Synthesis of compounds 10 and 11
Compounds 10 and 11 were synthesized as shown in Scheme 3. The starting 3-amino-2-cyclopropylamino-4-methylpyridine (27) was prepared from commercially available 2-hydroxy-4-methyl-3-nitropyridine (24) through a sequence involving treatment with POCl3, followed by chloro displacement from the resulting 2-chloro compound with the aminocyclopropyl group, and finally reducing the nitro to the amino group. 2-Chloro-5-nitronicotinic acid (30) was prepared by nitration of commercially available 2-hydroxynicotinic acid (28) followed by treatment with POCl3. Then amine 27 and the acid 30 underwent coupling to produce carboxamide 31. Diazepinone ring closure was performed by heating 31 in hexamethyldisilazane. Afterwards, the nitro group was reduced to produce the hydrochloride salt 10. Treatment of 10 with 50% aqueous NaOH yielded its corresponding free amino compound 11.
Scheme 3: Reagents and conditions: (a) POCl3, 150 °C, 6 h, 85%; (b) cyclopropylamine, xylene, 105 °C, 4 h, 99%; (c) SnCl2·2H2O, conc. HCl, CH3COOH, rt, 3 h, 83%; (d) 69% HNO3, conc. H2SO4, 50 °C, 7 h, 79%; (e) POCl3, reflux, 4 h, 78%; (f) i) 30, (COCl)2, benzene, DMF, rt, 1 h; ii) 27, THF, DIPEA, rt, 5 h, 53%; (g) HMDS, 110 °C, 24 h, 90%; (h) SnCl2·2H2O, conc. HCl, CH3COOH, rt, 3h, 73%; (i) 50% aq. NaOH, rt, 1 h, 90%.
Scheme 3: Reagents and conditions: (a) POCl3, 150 °C, 6 h, 85%; (b) cyclopropylamine, xylene, 105 °C, 4 h, 99...
Biological testing against HIV-1 reverse transcriptase
The results from the biological testing of all compounds synthesized, compared with nevirapine (1) and 9, against the wild-type RT together with K103N and Y181C mutant RT are shown in Table 1. It was found that compounds 2–8 exhibited higher inhibitory activity against WT-RT and both mutant RTs compared to nevirapine. Interestingly, 5 and 6 were found to be about four times more potent against WT-RT than 9, and they provided comparable activity against K103N mutant RT. Incorporation of a methyl group at the R1 position and the free N of the amide seems to be responsible for this higher activity. However, 9 showed better potency against the Y181C mutant RT compared to the other two compounds. Compound 5, without methoxy substituent, was found to be slightly more potent than 6 except for Y181C mutant RT. Additional N-methyl groups in 7 and 8 led to diminished activity relative to that of 5 and 6. 10 and 11, 8-amino analogues of nevirapine, were found to be ineffective inhibitors.
Table 1: Inhibitory activity of the synthesized compounds against HIV-1 reverse transcriptase.
| Compounds | IC50a (μM) | ||
|---|---|---|---|
| WT | K103N | Y181C | |
| nevirapine (1) | 1.070 ± 0.60b | 27.10 ± 5.20 | 228.5 ± 24.84 |
| 2 | 0.427 ± 0.31 | 6.23 ± 2.48 | 1.50 ± 0.34 |
| 3 | 0.757 ± 0.15 | 19.40 ± 2.80 | 2.90 ± 0.19 |
| 4 | 0.183 ± 0.10 | 13.90 ± 1.23 | 0.459 ± 0.29 |
| 5 | 0.0186 ± 0.002 | 0.224 ± 0.14 | 0.269 ± 0.08 |
| 6 | 0.0229 ± 0.01 | 0.428 ± 0.39 | 0.0593 ± 0.07 |
| 7 | 0.124 ± 0.03 | 4.37 ± 0.66 | 0.507 ± 0.36 |
| 8 | 0.0828 ± 0.03 | 4.59 ± 2.23 | 0.118 ± 0.06 |
| 9 | 0.0858 ± 0.00001 | 0.39 ± 0.23 | 0.00463 ± 0.0009 |
| 10 | 17.40 ± 2.11 | 62.10 ± 5.14 | 126.0 ± 28.03 |
| 11 | 6.05 ± 1.60 | 97.0 ± 16.8 | 61.0 ± 6.9 |
aIC50 is the concentration of inhibitor required for 50% inhibition of reverse transcriptase. bStandard errors obtained from duplicate experiments.
Molecular docking
To understand the binding mode of the new potent derivatives 5, 6 and 9 were docked into the HIV-1 RT binding site by using the default parameters of the GOLD v3.2 program. The wild type HIV-1 RT structure (pdb code 1klm) was taken from the protein data bank. Additionally, two HIV-1 RT mutants, K103N and Y181C, were used and analyzed by mutating positions 103 and 181 of the wild-type structure through the use of the Sybyl 7.2 program. The docked conformations of 5, 6, 9 and nevirapine are shown in Figure 3, and their GoldScores are presented in Table 2. In the wild-type and K103N binding sites, the docked orientations of 5, 6 and 9 are similar to that of nevirapine. In the Y181C binding site, except for 5, the orientations of the others were similar to nevirapine orientation. The GoldScores of 5, 6 and 9 were higher than those of nevirapine by 17.61–24.56, 9.52–15.00 and 15.33–21.06 in the WT, K103N, and Y181C binding pockets, respectively. In the wild-type binding pocket, the H-bond interaction with the backbone oxygen atom of Lys103 was found to contain 5, 6 and 9 but no nevirapine was present. Compounds 5, 6 and 9 also formed an H-bond interaction with the backbone nitrogen atom of Val106. Since there is a methyl group at the R1 position of 5 and 6, their docked conformations were slightly shifted below the binding pocket as compared to the docked conformation of 9. This shift caused the formation of a stronger H-bond interaction of 5 and 6 with Lys101, Val179, Tyr188 and Val189 compared to 9. The methyl group at R1 position of 5 and 6 also formed stronger H–π interaction with Trp229. Moreover, the methoxy substituent of 6 revealed a strong attractive interaction with Lys104, but the movement of ring C in 6 caused a steric interaction with the side chain of Lys102.
Figure 3: Docked orientations of nevirapine (green), 9 (yellow), 5, and 6 (atom type color – carbon: grey, chloride: green, hydrogen: white, nitrogen: blue, oxygen: red and sulfur: yellow) in WT (a), K103N (b), and Y181C (c) binding pockets.
Figure 3: Docked orientations of nevirapine (green), 9 (yellow), 5, and 6 (atom type color – carbon: grey, ch...
Table 2: GoldScores of nevirapine, 5, 6 and 9 in wild-type (WT), K103N and Y181C HIV-1 RT.
| Compounds | GoldScores | ||
|---|---|---|---|
| WT | K103N | Y181C | |
| nevirapine | 58.13 (± 0.27)a | 58.66 (± 0.39) | 56.92 (± 0.04) |
| 5 | 72.07 (± 1.60) | 79.23 (± 1.42) | 66.44 (± 0.63) |
| 6 | 79.19 (± 0.83) | 83.22 (± 0.85) | 71.92 (± 1.69) |
| 9 | 73.66 (± 2.36) | 76.27 (± 1.00) | 68.84 (± 1.09) |
aIn parenthesis is the standard error of the GoldScore from the triplicate of docking calculations.
In the case of the docked conformations of 5, 6 and 9 in the K103N binding pocket, the H-bond interactions with the backbone atom of Asn103 were still detected, but their H-bond interactions with Val106 were lost. Furthermore, the docked conformation of 5 showed stronger H-bond interaction with Lys101 compared to 6 and 9. The adjustment of the ethyl group also formed an important H-bond interaction with the oxygen atom of carbonyl Val179 at distances of 2.49, 2.74 and 2.68 Å for 5, 6 and 9 respectively. The methyl group at the R1 position of 5 and 6 presented the H–π interaction with side chain Trp229 closer than the hydrogen atom of 9.
In the Y181C binding pocket, it was found that the docked conformation of 5 had a different orientation compared to 6 and 9. Due to this orientation change of 5, some attractive interactions found in the wild-type binding pocket were lost. It was observed that the docked conformations of 6 and 9 were aligned in the same orientation as nevirapine. For 9, stronger attractive interactions with the backbone oxygen atoms of Val179 and Tyr188 in the Y181C binding pocket were formed as compared with the WT binding pocket. For 6, H-bond interaction between a hydrogen atom of the methoxy group of 6 and an oxygen atom of backbone Lys104 revealed a longer bond length (3.58 Å) in the Y181C binding pocket as compared to the WT binding pocket (2.14 Å).
Conclusion
The remarkable anti HIV-1 activity of dipyridodiazepinone derivatives, particularly compounds 5 and 6, was presented in this study. A preliminary SAR study showed that the methyl group at the R1 position and the free N of amide are crucial for potent activity. This is possibly because of the strong interaction with the amino acid residue in the RT enzyme. The secondary test of these two compounds was regarded to be valuable for future investigation.
Supporting Information
Supporting information provides details about the chemical methods, analytical data and biological testing.
| Supporting Information File 1: Experimental part. | ||
| Format: DOC | Size: 113.5 KB | Download |
Acknowledgements
We would like to thank the Thailand Research Fund (DBG4780009 for S.T., RTA5080005 for S.H., DBG5180022 for P.P., and MRG5080267 for P.S.), National Synchrotron Research Center for K.C., and the Ministry of Education for their financial support. We also gratefully acknowledge the help of Dr. Carrie Dykes of the University of Rochester, New York for the mutant clones, K103N and Y181C; and Dr. Hiroaki Mitsuya of the HIV and AIDS Malignancy Branch, USA for the WT clone.
References
-
Hargrave, K. D.; Proudfoot, J. R.; Grozinger, K. G.; Cullen, E.; Kapadia, S. R.; Patel, U. R.; Fuchs, V. U.; Mauldin, S. C.; Vitous, J.; Behnke, M. L.; Klunder, J. M.; Pal, K.; Skiles, J. W.; McNeil, D. W.; Rose, J. M.; Chow, G. C.; Skoog, M. T.; Wu, J. C.; Schmidt, G.; Engel, W. W.; Eberlein, W. G.; Saboe, T. D.; Campbell, S. J.; Rosenthal, A. S.; Adams, J. J. Med. Chem. 1991, 34, 2231–2241. doi:10.1021/jm00111a045
Return to citation in text: [1] -
O’Meara, J. A.; Yoakim, C.; Bonneau, P. R.; Bös, M.; Cordingley, M. G.; Déziel, R.; Doyon, L.; Duan, J.; Garneau, M.; Guse, I.; Landry, S.; Malenfant, E.; Naud, J.; Ogilvie, W. W.; Thavonekham, B.; Simoneau, B. J. Med. Chem. 2005, 48, 5580–5588. doi:10.1021/jm050255t
Return to citation in text: [1] -
Yoakim, C.; Bonneau, P. R.; Déziel, R.; Doyon, L.; Duan, J.; Guse, I.; Landry, S.; Malenfant, E.; Naud, J.; Ogilvie, W. W.; O’Meara, J. A.; Plante, R.; Simoneau, B.; Thavonekham, B.; Bös, M.; Cordingley, M. G. Bioorg. Med. Chem. Lett. 2004, 14, 739–742. doi:10.1016/j.bmcl.2003.11.049
Return to citation in text: [1] -
Cywin, C. L.; Klunder, J. M.; Hoermann, M.; Brickwood, J. R.; David, E.; Grob, P. M.; Schwartz, R.; Pauletti, D.; Barringer, K. J.; Shih, C.-K.; Sorge, C. L.; Erickson, D. A.; Joseph, D. P.; Hattox, S. E. J. Med. Chem. 1998, 41, 2972–2984. doi:10.1021/jm9707030
Return to citation in text: [1] [2] [3] -
Klunder, J. M.; Hoermann, M.; Cywin, C. L.; David, E.; Brickwood, J. R.; Schwartz, R.; Barringer, K. J.; Pauletti, D.; Shih, C.-K.; Erickson, D. A.; Sorge, C. L.; Joseph, D. P.; Hattox, S. E.; Adams, J.; Grob, P. M. J. Med. Chem. 1998, 41, 2960–2971. doi:10.1021/jm9707028
Return to citation in text: [1] -
Kelly, T. A.; McNeil, D. W.; Rose, J. M.; David, E.; Shih, C.-K.; Grob, P. M. J. Med. Chem. 1997, 40, 2430–2433. doi:10.1021/jm960837y
Return to citation in text: [1] -
Kelly, T. A.; Proudfoot, J. R.; McNeil, D. W.; Patel, U. R.; David, E.; Hargrave, K. D.; Grob, P. M.; Cardozo, M.; Agarwal, A.; Adams, J. J. Med. Chem. 1995, 38, 4839–4847. doi:10.1021/jm00024a011
Return to citation in text: [1] -
Proudfoot, J. R.; Hargrave, K. D.; Kapadia, S. R.; Patel, U. R.; Grozinger, K. G.; McNeil, D. W.; Cullen, E.; Cardozo, M.; Tong, L.; Kelly, T. A.; Rose, J. M.; David, E.; Mauldin, S. C.; Fuchs, V. U.; Vitous, J.; Hoermann, M.; Klunder, J. M.; Raghavan, P.; Skiles, J. W.; Mui, P.; Richman, D. D.; Sullivan, J. L.; Shih, C.-K.; Grob, P. M.; Adams, J. J. Med. Chem. 1995, 38, 4830–4838. doi:10.1021/jm00024a010
Return to citation in text: [1] -
Khunnawutmanotham, N.; Chimnoi, N.; Saparpakorn, P.; Pungpo, P.; Louisirirotchanakul, S.; Hannongbua, S.; Techasakul, S. Molecules 2007, 12, 218–230. doi:10.3390/12020218
Return to citation in text: [1] [2] -
Grozinger, K. G.; Fuchs, V. U.; Hargrave, K. D.; Mauldin, S.; Vitous, J.; Campbell, S.; Adams, J. J. Heterocycl. Chem. 1995, 32, 259–263. doi:10.1002/jhet.5570320144
Return to citation in text: [1] -
Bobbitt, J. M.; Scola, D. J. Org. Chem. 1960, 25, 560–564. doi:10.1021/jo01074a018
Return to citation in text: [1]
| 1. | Hargrave, K. D.; Proudfoot, J. R.; Grozinger, K. G.; Cullen, E.; Kapadia, S. R.; Patel, U. R.; Fuchs, V. U.; Mauldin, S. C.; Vitous, J.; Behnke, M. L.; Klunder, J. M.; Pal, K.; Skiles, J. W.; McNeil, D. W.; Rose, J. M.; Chow, G. C.; Skoog, M. T.; Wu, J. C.; Schmidt, G.; Engel, W. W.; Eberlein, W. G.; Saboe, T. D.; Campbell, S. J.; Rosenthal, A. S.; Adams, J. J. Med. Chem. 1991, 34, 2231–2241. doi:10.1021/jm00111a045 |
| 9. | Khunnawutmanotham, N.; Chimnoi, N.; Saparpakorn, P.; Pungpo, P.; Louisirirotchanakul, S.; Hannongbua, S.; Techasakul, S. Molecules 2007, 12, 218–230. doi:10.3390/12020218 |
| 9. | Khunnawutmanotham, N.; Chimnoi, N.; Saparpakorn, P.; Pungpo, P.; Louisirirotchanakul, S.; Hannongbua, S.; Techasakul, S. Molecules 2007, 12, 218–230. doi:10.3390/12020218 |
| 4. | Cywin, C. L.; Klunder, J. M.; Hoermann, M.; Brickwood, J. R.; David, E.; Grob, P. M.; Schwartz, R.; Pauletti, D.; Barringer, K. J.; Shih, C.-K.; Sorge, C. L.; Erickson, D. A.; Joseph, D. P.; Hattox, S. E. J. Med. Chem. 1998, 41, 2972–2984. doi:10.1021/jm9707030 |
| 2. | O’Meara, J. A.; Yoakim, C.; Bonneau, P. R.; Bös, M.; Cordingley, M. G.; Déziel, R.; Doyon, L.; Duan, J.; Garneau, M.; Guse, I.; Landry, S.; Malenfant, E.; Naud, J.; Ogilvie, W. W.; Thavonekham, B.; Simoneau, B. J. Med. Chem. 2005, 48, 5580–5588. doi:10.1021/jm050255t |
| 3. | Yoakim, C.; Bonneau, P. R.; Déziel, R.; Doyon, L.; Duan, J.; Guse, I.; Landry, S.; Malenfant, E.; Naud, J.; Ogilvie, W. W.; O’Meara, J. A.; Plante, R.; Simoneau, B.; Thavonekham, B.; Bös, M.; Cordingley, M. G. Bioorg. Med. Chem. Lett. 2004, 14, 739–742. doi:10.1016/j.bmcl.2003.11.049 |
| 4. | Cywin, C. L.; Klunder, J. M.; Hoermann, M.; Brickwood, J. R.; David, E.; Grob, P. M.; Schwartz, R.; Pauletti, D.; Barringer, K. J.; Shih, C.-K.; Sorge, C. L.; Erickson, D. A.; Joseph, D. P.; Hattox, S. E. J. Med. Chem. 1998, 41, 2972–2984. doi:10.1021/jm9707030 |
| 5. | Klunder, J. M.; Hoermann, M.; Cywin, C. L.; David, E.; Brickwood, J. R.; Schwartz, R.; Barringer, K. J.; Pauletti, D.; Shih, C.-K.; Erickson, D. A.; Sorge, C. L.; Joseph, D. P.; Hattox, S. E.; Adams, J.; Grob, P. M. J. Med. Chem. 1998, 41, 2960–2971. doi:10.1021/jm9707028 |
| 6. | Kelly, T. A.; McNeil, D. W.; Rose, J. M.; David, E.; Shih, C.-K.; Grob, P. M. J. Med. Chem. 1997, 40, 2430–2433. doi:10.1021/jm960837y |
| 7. | Kelly, T. A.; Proudfoot, J. R.; McNeil, D. W.; Patel, U. R.; David, E.; Hargrave, K. D.; Grob, P. M.; Cardozo, M.; Agarwal, A.; Adams, J. J. Med. Chem. 1995, 38, 4839–4847. doi:10.1021/jm00024a011 |
| 8. | Proudfoot, J. R.; Hargrave, K. D.; Kapadia, S. R.; Patel, U. R.; Grozinger, K. G.; McNeil, D. W.; Cullen, E.; Cardozo, M.; Tong, L.; Kelly, T. A.; Rose, J. M.; David, E.; Mauldin, S. C.; Fuchs, V. U.; Vitous, J.; Hoermann, M.; Klunder, J. M.; Raghavan, P.; Skiles, J. W.; Mui, P.; Richman, D. D.; Sullivan, J. L.; Shih, C.-K.; Grob, P. M.; Adams, J. J. Med. Chem. 1995, 38, 4830–4838. doi:10.1021/jm00024a010 |
| 4. | Cywin, C. L.; Klunder, J. M.; Hoermann, M.; Brickwood, J. R.; David, E.; Grob, P. M.; Schwartz, R.; Pauletti, D.; Barringer, K. J.; Shih, C.-K.; Sorge, C. L.; Erickson, D. A.; Joseph, D. P.; Hattox, S. E. J. Med. Chem. 1998, 41, 2972–2984. doi:10.1021/jm9707030 |
| 10. | Grozinger, K. G.; Fuchs, V. U.; Hargrave, K. D.; Mauldin, S.; Vitous, J.; Campbell, S.; Adams, J. J. Heterocycl. Chem. 1995, 32, 259–263. doi:10.1002/jhet.5570320144 |
| 11. | Bobbitt, J. M.; Scola, D. J. Org. Chem. 1960, 25, 560–564. doi:10.1021/jo01074a018 |
© 2009 Khunnawutmanotham et al; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (http://www.beilstein-journals.org/bjoc)