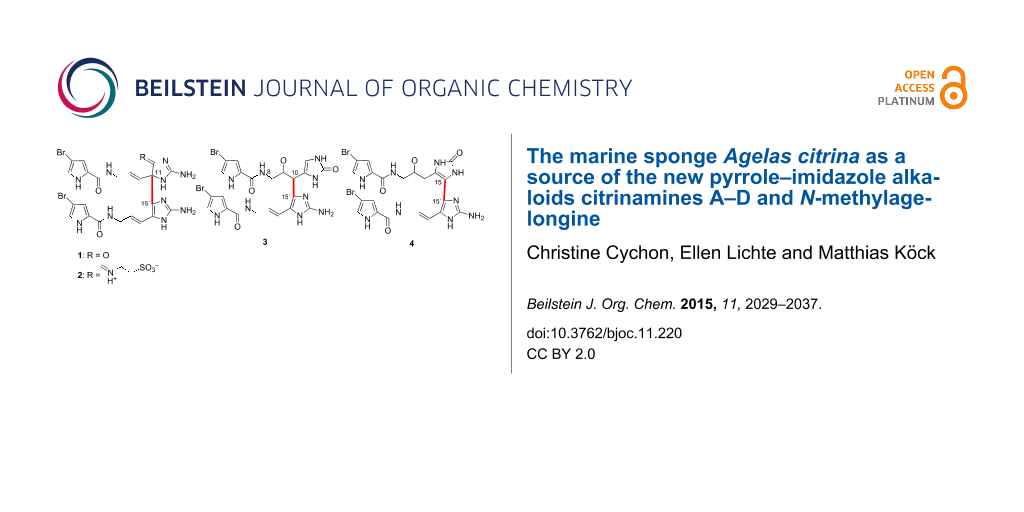
Abstract
The chemical investigation of the Caribbean sponge Agelas citrina revealed four new pyrrole–imidazole alkaloids (PIAs), the citrinamines A–D (1–4) and the bromopyrrole alkaloid N-methylagelongine (5). All citrinamines are dimers of hymenidin (6) which was also isolated from this sponge as the major metabolite. Citrinamines A (1) and B (2) are derivatives of the PIA dimer mauritiamine (7), whereas citrinamine C (3) is derived from the PIA dimer nagelamide B (8). Citrinamine D (4) shows an uncommon linkage between the imidazole rings of both monomeric units as it is only observed in the benzocyclobutane ring moiety of benzosceptrins A–C (9–11). Compound 5 is the N-methyl derivative of agelongine (12) which consist of a pyridinium ring and an ester linkage instead of the aminoimidazole moiety and the common amide bond in PIAs.
Graphical Abstract
Introduction
The family of pyrrole-imidazole alkaloids (PIAs) represents a fascinating example of a large variety of secondary metabolites produced exclusively by marine sponges. To date, more than 150 PIAs have been isolated mainly from various species of the families Agelasidae, Axinellidae, Dyctionellidae, and Hymeniacidonidae and some of them show promising biological activities [1]. The chemical investigation of the Caribbean sponge Agelas citrina yielded the following known monomeric PIAs: hymenidin (6) [2], keramadine (13) [3], dispacamide B (14) [4], mukanadin B (15) [5], 2-debromotaurodispacamide A (16) [6], tauroacidin B (17) [7], and the dimeric PIA benzosceptrin B (10) [8-10] (Figure 1). Additionally, five new compounds, four with a dimeric PIA structure (1–4) and the N-methyl analogue (5) of the bromopyrrole alkaloid agelongine (12) [11], were isolated. Herein, we describe the isolation and structure elucidation of citrinamines A (1), B (2), and N-methylagelongine (5). The identification of citrinamines C (3) and D (4), which were both only obtained as mixtures, will also be discussed in this context.
Figure 1: Selected pyrrole-imidazole alkaloids (1–4, 6–11, and 13–20), and agelongine analogues (5, 12, and 21).
Figure 1: Selected pyrrole-imidazole alkaloids (1–4, 6–11, and 13–20), and agelongine analogues (5, 12, and 21...
Results and Discussion
The crude extract of the sponge Agelas citrina (collected in the Bahamas in March 2001) was investigated by a standard separation scheme. The MeOH/CH2Cl2 extract of the sample was partitioned between n-hexane, n-BuOH, and H2O. The n-BuOH soluble fraction was further purified by size exclusion chromatography (Sephadex LH-20) and preparative reversed-phase HPLC yielding five new compounds (1–5). The isolation and structure elucidation of compounds 1 to 5 are discussed in detail.
The molecular weight of citrinamine A (1) was obtained from ESI mass spectrometry (HR–ESIMS, m/z 633.0330, [M + H]+, monoisotopic), together with the pseudomolecular ion peaks at m/z = 633/635/637 (1:2:1) the molecular formula C22H23Br2N10O3 was derived. The structure of 1 is very similar to mauritiamine (7) [12] and its 2´-debromo derivative nagelamide P (18) [13]. The 1D and 2D 1H and 13C NMR spectra of 1 showed two additional signals for sp2 methines indicating two 3-bromopyrrole carboxamide moieties (Table 1). The similarities of the NMR data of 1 with those of mauritiamine (7) and nagelamide P (18) proved the same connectivity of the respective monomers hymendin (6) and/or oroidin (19) (Figure 2). The linkage of both monomeric units between the sp3 quaternary C-11 (65.0 ppm) and the sp2 quaternary C-15´ (117.4 ppm) is indicated by the 1H,1H-NOESY [H-9 (5.87 ppm) to H-10´ (6.47 ppm) and H-10 (5.81 ppm) to H-10´] and the 1H,13C-HMBC correlations [H-9 to C-11, H-10 to C-11, H-9´ (6.10 ppm) to C-11´ (122.5 ppm), and H-10´ to C-11´] as well as by the absence of the H-15´ signal in the 1D 1H NMR spectrum. The missing signal for H-15 in the 1D 1H NMR spectrum and a sp2 quaternary carbon (δC 171.7 ppm) in the 1D 13C spectrum verified the oxidation of one aminoimidazole ring at position C-15 like in mauritiamine (7) and nagelamide P (18).
Table 1: 1H, 13C, and 15N chemical shifts of citrinamine A (1) (600 and 850 MHz, DMSO-d6) in comparison to mauritiamine (7) (600 MHz, DMSO-d6) and nagelamide P (18) (500 MHz, DMSO-d6).
| Pos. | citrinamine A (1)a | mauritiamine (7)a | nagelamide P (18) [13] | |||
|---|---|---|---|---|---|---|
| δH, mult. (J/Hz) | δC/δN | δH, mult. (J/Hz) | δC/δN | δH, mult. (J/Hz) | δC/δN | |
| 1-NH | 11.80, s | (161) | 12.66, s | 12.68, brs | ||
| 2 | 7.02, dd (1.5; 2.8) | 121.1 | – | 104.6 | – | 104.6 |
| 3 | – | 95.0 | – | 97.9 | – | 98.0 |
| 4 | 6.87b | 111.3 | 6.95, brs | 112.8 | 6.95, brs | 112.8 |
| 5 | – | 127.0 | – | 127.9 | – | 128.4 |
| 6 | – | 159.5 | – | 158.9 | – | 159.5 |
| 7-NH | 8.39, t (5.9) | (104) | 8.39, t (6.1) | 8.42, brt (5.9) | ||
| 8 | 3.90, dd (4.8; 9.6) | 39.1 | 3.88, m | 39.2 | 3.87, brt (5.3) | 40.6 |
| 9 | 5.87, m | 130.6 | 5.82b | 128.9 | 6.02, dt, (16.0; 5.3) | 139.1 |
| 10 | 5.81, d (15.8) | 125.4 | 5.81b | 126.4 | 6.05, brd | 126.7 |
| 11 | – | 65.0 | – | 65.4 | – | 69.9 |
| 12-NH | (101) | |||||
| 13 | – | 147.5 | – | 148.0 | – | 147.7 |
| 13-NH2 | 7.62, brs | |||||
| 14-NH | 12.12, brs | |||||
| 15 | – | 171.7 | – | 175.0 | – | 183.4 |
| 1´-NH | 11.80, s | (161) | 12.66, s | 11.80, brs | ||
| 2´ | 6.99, dd (1.4; 2.8) | 121.1 | – | 104.6 | 6.98, brs | 121.4 |
| 3´ | – | 95.0 | – | 97.9 | – | 95.0 |
| 4´ | 6.89b | 111.3 | 6.85, brs | 112.8 | 6.86, brs | 111.7 |
| 5´ | – | 127.0 | – | 127.9 | – | 126.6 |
| 6´ | – | 159.4 | – | 158.7 | – | 158.8 |
| 7´-NH | 8.41, t (5.8) | (107) | 8.42, t (5.7) | 8.39, brt (5.9) | ||
| 8´ | 4.01, m | 40.1 | 3.97, m | 40.1 | 3.95, m | 40.5 |
| 9´ | 6.10, m | 129.0 | 6.05, m | 128.3 | 6.02, dt (16.0; 5.3) | 136.6 |
| 10´ | 6.47, d (15.8) | 116.8 | 6.48 | 116.4 | 6.41, brd (16.0) | 115.7 |
| 11´ | – | 122.5 | – | 121.4 | – | 119.2 |
| 12´-NH | (132) | 12.56, brs | ||||
| 13´ | – | 147.5 | – | 148.0 | – | 147.7 |
| 13´-NH2 | 7.62, brs | |||||
| 14´-NH | ||||||
| 15´ | – | 117.4 | – | 117.5 | – | 121.3 |
a1H and 13C chemical shifts are referenced to the DMSO-d6 signal (2.50 ppm and 39.5 ppm, respectively). 15N NMR shifts were not calibrated with an external standard. Therefore, the δ value has an accuracy of about 1 ppm in reference to NH3 (0 ppm) and the 15N NMR shifts are given without decimals. bNo multiplicity information could be given because of overlapped signals.
![[1860-5397-11-220-2]](/bjoc/content/figures/1860-5397-11-220-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red arrows) correlations for citrinamine A (1).
Figure 2: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red...
Based on 1H,1H-NOESY correlations H-8 (3.90 ppm) to H-10 and H-8´ (4.01 ppm) to H-10´ and the corresponding coupling constants for H-9/H-10 and H-9´/H-10´ (both 15.8 Hz), both of the two double bonds C-9 (130.6 ppm) and C-10 (125.4 ppm) as well as C-9’ (129.0 ppm) and C-10’ (116.8 ppm) were assigned to the E configuration. The same double bond geometries were also observed for mauritiamine (7) and nagelamide P (18). Citrinamine A (1) is the 2,2´-didebromo derivative of mauritiamine (7) and as in the original publications of 7 and 18 no chiroptical effect was observed for 1. Synthetic studies on mauritiamine (7) [14] demonstrated the formation of similar racemic products by a chemical oxidative dimerization which could be an alternative origin of these metabolites.
The molecular formula of citrinamine B (2) was established by HR–ESIMS (m/z 740.0337, [M + H]+, monoisotopic) and the pseudomolecular ion peaks at m/z = 740/742/744 (1:2:1) to be C24H28Br2N11O5S. The 1D 1H and 13C NMR spectra of 2 were similar to those observed for 1, except for the additional signals of one amine and two methylene groups (Table 2). The analysis of the 1H,1H-COSY and the 1H,13C-HMBC spectra linked these new signals to an aminoethyl chain and the comparison with the molecular formula indicated the existence of a taurine moiety in compound 2. This structural proposal for citrinamine B (2) was proven by the 1H,13C-HMBC correlations H-2´´ (3.69/3.55 ppm) to C-3´´ (48.6 ppm) and C-15 (178.0 ppm), the 1H,15N-HMBC correlations H-3´´ (2.89/2.80 ppm) to N-1´´ (116 ppm), and the 1H,1H-NOESY correlation H-9/10 (5.95 ppm) to H-1´´ (9.85 ppm) (Figure 3).
Table 2: 1H, 13C, and 15N chemical shifts of citrinamine B (2) (600 and 850 MHz, DMSO-d6) in comparison to nagelamide H (20) (600 MHz, DMSO-d6).
| Pos. | citrinamine B (2)a | nagelamide H (20) [15] | ||
|---|---|---|---|---|
| δH, mult. (J/Hz) | δC/δN | δH, mult. (J/Hz) | δC/δN | |
| 1-NH | 11.81, s | (161) | 11.71, s | |
| 2 | 7.05, dd (1.4; 2.8) | 121.7 | – | 104.8 |
| 3 | – | 95.4 | – | 97.9 |
| 4 | 6.90, dd (1.7; 2.4) | 111.8 | 7.00, s | 113.0 |
| 5 | – | 126.8 | – | 127.8 |
| 6 | – | 160.0 | – | 158.8 |
| 7-NH | 8.44, t (5.7) | (104) | 8.12, t (5.9) | |
| 8 | 3.92b | 39.7 | 3.86, mc; 4.04, mc | 40.1c |
| 9 | 5.95b | 130.6 | 6.02, dt (6.0; 15.2)c | 124.7c |
| 10 | 5.95b | 124.4 | 6.15, d (15.2)c | 115.4c |
| 11 | – | 68.0 | – | 112.8c |
| 12-NH | ||||
| 13 | – | 167.7 | – | 167.3 |
| 13-NH2 | 9.10, s; 8.59, s | (90) | 8.79, brs; 9.11, brs. | |
| 14-NH | 10.04, s | (109) | 10.19, brs | |
| 15 | – | 178.0 | – | 177.4 |
| 1´-NH | 11.80, s | (161) | 12.72, s | |
| 2´ | 7.01, dd (1.4; 2.8) | 121.7 | – | 104.8 |
| 3´ | – | 95.2 | – | 98.0 |
| 4´ | 6.93, dd (1.7; 2.3) | 111.8 | 6.97, s | 113.0 |
| 5´ | – | 126.8 | – | 127.9 |
| 6´ | – | 159.8 | – | 158.8 |
| 7´-NH | 8.48, t (5.8) | (106) | 8.21, t (5.9) | |
| 8´ | 4.15, m; 3.91b | 39.9 | 3.92, mc | 39.3c |
| 9´ | 6.14, m | 130.7 | 5.99, dt (6.2; 15.3)c | 130.8c |
| 10´ | 6.26, d (16.1) | 115.2 | 5.90, d (15.3)c | 129.6c |
| 11´ | – | 124.5 | – | 69.7c |
| 12´-NH | (132) | 12.60, brs | ||
| 13´ | – | 147.7 | – | 148.3 |
| 13´-NH2 | 7.74, brs | |||
| 14´-NH | 13.02, brs | |||
| 15´ | – | 117.5 | – | 123.1 |
| 1´´-NH | 9.85, s | (116) | 9.89, brs | |
| 2´´ | 3.69, m; 3.55, m | 41.0 | 3.65, m; 3.65, m | 40.3 |
| 3´´ | 2.89, m; 2.80, m | 48.6 | 2.81, t (7.1) | 48.2 |
a1H and 13C chemical shifts are referenced to the DMSO-d6 signal (2.50 ppm and 39.5 ppm respectively). 15N NMR shifts were not calibrated with an external standard. Therefore, the δ value has an accuracy of about 1 ppm in reference to NH3 (0 ppm) and the 15N NMR shifts are given without decimals. bNo multiplicity information could be given because of overlapped signals. cPossible assignment error, see text for details.
![[1860-5397-11-220-3]](/bjoc/content/figures/1860-5397-11-220-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red arrows) correlations for citrinamine B (2).
Figure 3: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red...
The structure of 2 is very similar to nagelamide H (20) [15] which is a mauritiamine derivative with a taurine residue in position C-15. There is probably an assignment error of the carbons C-8 to C-11 and C-8´ to C-11´ in the original publication of nagelamide H (20). All mauritiamine derivatives have comparable chemical shifts of these moieties and therefore a mixing up is plausible.
Based on NOESY correlations H-8 (3.92 ppm) to H-9/10 and H-8´ (4.15/3.91 ppm) to H-10´ (6.26 ppm) as well as the corresponding 3JHH coupling constant H-9´ (6.14 ppm)/H-10´ (16.1 Hz), both of the double bonds of 2 were assigned to E configuration. Citrinamine B (2) is the 2,2´-didebromo derivative of nagelamide H (20). We could not observe a chiroptical effect for 2 as it was also described in the original work of 20.
Our investigation on Agelas citrina yielded two additional pyrrole–imidazole alkaloids, citrinamines C (3) and D (4), which were both obtained as mixtures. The isolation of the pure compounds of 3 and 4 by preparative chromatography failed but the analysis of the mixtures allowed the identification of their structures. The molecular weights of citrinamine C (3) (m/z 650.0493, [M + H]+, monoisotopic) and citrinamine D (4) (m/z 650.0470, [M + H]+, monoisotopic), and the respective pseudo-molecular ion peaks at m/z = 650/652/654 (1:2:1) indicated the molecular formula C23H26Br2N9O4 for both compounds. The analysis of the 1D and 2D 1H and 13C NMR spectra revealed for both compounds a dimeric hymenidin structure (Figure 4, Figure 5 and Supporting Information File 1, Table S1).
![[1860-5397-11-220-4]](/bjoc/content/figures/1860-5397-11-220-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), and 1H,13C-HMBC (red arrows) correlations for citrinamine C (3).
Figure 4: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), and 1H,13C-HMBC (red arrows) correlati...
![[1860-5397-11-220-5]](/bjoc/content/figures/1860-5397-11-220-5.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 5: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red arrows) correlations for citrinamine D (4).
Figure 5: Selected 1H,1H-COSY (blue bonds), 1H,1H-NOESY (blue arrows), 1H,13C-HMBC, and 1H,15N-HMBC (both red...
The 1H,13C-HMBC correlations of citrinamine C (3) from H-10 (4.09 ppm) to C-11´ (121.3 ppm) and C-15´ (117.9 ppm), suggested a connection of C-10 (34.5 ppm) with the imidazole ring of the second subunit as it was also described for nagelamide B (8) [15]. The structure of citrinamine C (3) was elucidated to be the 2,2´-didebromo derivative of nagelamide B (8) with an additional methylation of the hydroxy group at C-9 (80.8 ppm) and an oxidation of the imidazole ring at position C-13 (154.6 ppm) (an urea instead of a guanidine moiety). The relative configuration of the stereogenic centers C-9 and C-10 was identical as described for nagelamide B (8).
In contrast to citrinamine C (3), a different connection of the monomeric units was found for citrinamine D (4). Furthermore, 3 and 4 have an additional methoxy group at C-9 (80.8 and 78.1 ppm, respectively) and an oxidized imidazole ring compared to 1. The 1H,13C-HSQC spectrum of 4 disclosed for C-10 (27.2 ppm) a methylene group (δH 2.34 ppm) and no hydrogen signal for C-15 (105.5 ppm). The missing proton signals for C-15 and C-15´ (115.8 ppm) and the 1H,1H-NOESY peaks H-10/H-9´ (6.16 ppm) and H-10/H-10´ (6.14 ppm) indicated a connection of the oxidized imidazole and the aminoimidazole ring in positions C-15 and C-15´. This uncommon linkage occurred so far only in the benzocyclobutane ring system of the benzosceptrins A–C (9–11). The incorporation of methoxy groups at carbon C-9 in citrinamines C (3) and D (4) may be attributed to the utilization of MeOH as solvent for the extraction of the sponge. To verify the structure elucidation of citrinamines C (3) and D (4), further extraction of sponge tissue of Agelas citrina is necessary to obtain pure material.
The exact molecular weight of compound 5 was determined as m/z 353.0135 ([M + H]+, monoisotopic) corresponding to the molecular formula C14H14Br2N2O4. The pseudomolecular ion peaks at m/z = 353/355 (1:1) proved the presence of one bromine atom in 5. The detailed analysis of the 1D 1H and 13C NMR spectra of 5 revealed only signals for the 3-bromopyrrole ring (two quaternary carbons at δC 121.6 ppm/94.1 ppm and two sp2 methines at δC 130.2 ppm/118.9 ppm) but no signals for the aminoimidazole moiety (Table 3). The 1H,13C-HMBC correlations from H-2 (7.30 ppm) to C-17 (36.5 ppm), H-17 (3.75 ppm) to C-2 (130.2 ppm), and H-17 to C-5 (121.6 ppm) indicated a methylation of the pyrrole nitrogen (Figure 6) which was proven by the 1H,15N-HMBC correlations H-2 to N-1 (159 ppm), H-4 (6.91 ppm) to N-1, and H-17 to N-1. The correlation between the methyl group H-17 and C-6 (158.6 ppm) suggested a linkage with the carboxylic acid in position C-5. The 1H,1H-COSY spectrum showed the connectivity of the methylene groups H-8 (4.65 ppm) and H-9 (5.07 ppm) and the 1H,13C-HMBC correlation H-8 to C-6 linked the ethylene group with the N-methyl-3-bromopyrrole carboxylic acid via an ester bond. The 1D 1H NMR spectrum of 5 disclosed four additional aromatic protons and 1H,1H-COSY peaks connected H-13 (8.95 ppm) with H-14 (8.25 ppm) and H-15 (9.26 ppm). The multiplet pattern of the four proton signals (s for H-11, d for H-14 and H-16, and dd for H-15) proved a meta di-substituted aromatic ring in the molecule. This is further proven by 1H,13C-HMBC correlations, such as H-11 (9.62 ppm) to C-12 (128.0 ppm), C-13 (145.7 ppm), and C-15 (146.6 ppm), H-13 and H-15 to C-11 (146.5 ppm), and H-14 to C-12.
Table 3: 1H, 13C, and 15N chemical shifts of N-methylagelongine (5) (600 MHz, DMSO-d6) in comparison to agelongine (12) (500 MHz, MeOH-d4) and daminin (21) (400 and 500 MHz, DMSO-d6).
| Pos. | N-methylagelongine (5)a | agelongine (12) [11] | daminin (21) [16] | |||
|---|---|---|---|---|---|---|
| δH, mult. (J/Hz) | δC/δN | δH, mult. (J/Hz) | δC/δN | δH, mult. (J/Hz) | δC/δN | |
| 1-N | – | (159) | 12.3, s | |||
| 2 | 7.30, d (1.9) | 130.2 | 7.05, d (1.5) | 125.6 | 7.05, d (0.9) | 124.6 |
| 3 | – | 94.1 | – | 94.1 | 6.16, d (1.9) | 109.5 |
| 4 | 6.91, d (1.9) | 118.9 | 6.89, d (1.5) | 118.5 | 6.78, t (1.9; 0.9) | 115.7 |
| 5 | – | 121.6 | – | 123.0 | – | 120.7 |
| 6 | – | 158.6 | – | 161.0 | – | 159.0 |
| 8 | 4.65, t (4.6) | 62.6 | 4.80, t (5.5) | 63.6 | 4.65, t (3.7) | 61.8 |
| 9 | 5.07, t (4.6) | 59.8 | 5.07, t (5.5) | 61.7 | 5.01, t (3.7) | 58.9 |
| 10-N | – | (209) | – | – | ||
| 11 | 9.62, s | 146.5 | 9.42, s | 147.4 | 9.40, s | 145.7 |
| 12 | – | 128.0 | – | 140.2 | – | 140.9 |
| 13 | 8.95, d (7.9) | 145.7 | 8.99, dt (7.7; 1.5) | 147.0 | 8.78, d (7.4) | 144.7 |
| 14 | 8.25, dd (7.9; 6.4) | 133.0 | 8.14, dd (7.7; 6.2) | 128.8 | 8.06, t (7.4; 6.5) | 126.6 |
| 15 | 9.26, d (6.4) | 146.6 | 9.05, dt (6.2; 1.5) | 146.6 | 9.04, t (6.5) | 144.0 |
| 16 | – | 163.2 | – | 167.0 | – | 161.6 |
| 17 | 3.75, s | 36.5 | – | – | – | – |
a1H and 13C chemical shifts are referenced to the DMSO-d6 signal (2.50 ppm and 39.5 ppm respectively). 15N NMR shifts were not calibrated with an external standard. Therefore, the δ value has an accuracy of about 1 ppm in reference to NH3 (0 ppm) and the 15N NMR shifts are given without decimals.
![[1860-5397-11-220-6]](/bjoc/content/figures/1860-5397-11-220-6.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 6: Selected 1H,1H-COSY (blue bonds), 1H,13C-HMBC, and 1H,15N-HMBC (both red arrows) correlations for N-methylagelongine (5).
Figure 6: Selected 1H,1H-COSY (blue bonds), 1H,13C-HMBC, and 1H,15N-HMBC (both red arrows) correlations for N...
The investigation of the 1H,15N-HMBC spectrum of 5 revealed the correlations H-8, H-11, H-14, and H-15 to N-10 (209 ppm) and suggested the nitrogen N-10 in the aromatic ring which is connected to the aliphatic chain at position C-9 (59.8 ppm). The correlations H-11 and H-15 to C-9 proved this substitution and the correlations H-11 and H-13 to C-16 (163.2 ppm) showed the second substituent to be a formic acid group. The structure of compound 5 was identified to be the N-methyl derivative of agelongine (12). Debromoagelongine (daminin (21) [16]) is the third compound in the agelongine family. All agelongine analogues were isolated from marine sponges.
The citrinamines A–D (1–4) were further evaluated for antimicrobial and cytotoxic activity. Classical agar diffusion assays were performed using the fungus Aspergillus niger, the yeast Saccharomyces cerevisiae as well as the Gram-negative bacterium Escherichia coli, and the Gram-positive bacterium Micrococcus luteus and Mycobacterium phlei as test organisms. In agar diffusion assays with Mycobacterium phlei considerable inhibition zones were observed for citrinamines B, C, and D (2–4), while there was also an activity of citrinamine C (3) against Micrococcus luteus. All compounds (1–4) showed no inhibition of cell proliferation of mouse fibroblasts.
Conclusion
The analysis of the marine sponge Agelas citrina revealed four new compounds of the pyrrole–imidazole alkaloid (PIA) family. Citrinamines A (1) and B (2) are closely related to mauritiamine (7) which can be seen as the most less complex dimeric PIA (the first published one) in which the monomeric units are only connected by one bond (C-11/C-15’). As it was already described for mauritiamine (7) and its congeners nagelamide P (18) and nagelamide H (20), compounds 1 and 2 were obtained as racemic mixtures. Citrinamines C (3) and D (4) show a different connection of the two monomeric hymenidin units (C-10/C-15’ and C-15/C-15’) compared to mauritiamine (7). Citrinamine C (3) is very closely related to nagelamide B (8) which are both hydroxylated at C-9 (methoxy in 3, hydroxy in 8). The C-15/C-15’ linkage between the imidazole rings of both monomers in citrinamine D (4) is uncommon. This connection was only known from the benzosceptrins A–C (9–11) in which the two imidazole rings are connected by a benzene ring (benzocyclobutane ring system) and not by a single bond as in citrinamine D (4). Although, 3 and 4 have different connectivities, both compounds were also obtained as racemic mixtures. Finally, the pyrrole–pyridinium alkaloid N-methylagelongine (5) was also isolated from Agelas citrina. Compound 5 is the N-methylated pyrrole derivative of agelongine (12). The debromo compound is known as daminin (21). The citrinamines A–D (1–4) were tested against several pathogenic bacteria, fungi, and cultures of mice fibroblasts. Only minor antimicrobial activities were obtained for citrinamines B–D (2–4) whereas no activities were found in the cytotoxicity assay for citrinamines A–D (1–4).
Experimental
General experimental procedures
1H, 13C, and 15N NMR spectra were conducted on Bruker Avance I 400 MHz, Bruker Avance II 600 MHz, and Bruker Avance III 850 MHz NMR spectrometers. All experiments were measured at 303 K in DMSO-d6 as solvent. The DQF-1H,1H-COSY, 1H,13C-HSQC, 1H,13C-HMBC, 1H,15N-HSQC, 1H,15N-HMBC, and 1H,1H-NOESY experiments were carried out using standard parameters. The mixing time for NOESY spectra was set to 200 ms, and the delay for the HMBC measurements was set to 80 ms. HPLC-MS analysis were performed with an Agilent 1100 HPLC systems and Bruker Daltonics micrOTOFLC. Analytical chromatography: Waters XTerra RP18 column (3.0 mm × 150 mm, 3.5 µm) with a MeCN/H2O/HCOOH gradient [0 min: 10% MeCN/90% HCOOH (0.1%); 30 min: 60% MeCN/40% HCOOH (0.1%) with a flow rate of 0.4 mL min−1]. Preparative chromatography: Prontosil Eurobond C18 column (20 mm × 250 mm, 5 µm) with a MeCN/TFA (0.1%) gradient. UV spectra were recorded during HPLC analysis with a DAD (Agilent).
Animal material
The marine sponge Agelas citrina was collected by SCUBA diving on March 11, 2001 at San Salvador in the Bahamas (27 m depth). The samples were immediately frozen after collection and kept at −20 °C until extraction. A voucher specimen was deposited under registration no. ZMA POR. 17278 at the Zoölogical Museum, University of Amsterdam (The Netherlands). Sponge identification was kindly conducted by W. H. de Weerdt and Dr. R. W. M. van Soest, Institute for Biodiversity and Ecosystem Dynamics, Zoölogical Museum, University of Amsterdam, The Netherlands (new address of RWMvS: Netherlands Centre for Biodiversity, Department Marine Zoology, Leiden, The Netherlands).
Extraction and isolation
As already described in [17] the freeze-dried sponge tissue of Agelas citrina (120 g) was crushed with a mill and extracted exhaustively at room temperature with a 1:1 mixture of MeOH/CH2Cl2. Part of the crude extract (40.84 g) was partitioned between n-hexane (4 × 600 mL) and MeOH (450 mL). After evaporating, the MeOH extract was then partitioned between n-BuOH (3 × 600 mL) and H2O (450 mL). The resulting n-BuOH phase (22.02 g) from the solvent partitioning scheme was purified by gel chromatography on Sephadex LH-20 (Pharmacia) using MeOH as mobile phase. The final purification of the isolated compounds was achieved by preparative RP18 HPLC on a Prontosil Eurobond C18 column (20 mm × 250 mm, 5 µm) applying a MeCN/TFA (0.1%) gradient to yield 1 (9.2 mg, 0.0077% of dry weight), 2 (4.0 mg, 0.0033%), 3 (1.7 mg, 0.0014%), 4 (5.1 mg, 0.0043%), 5 (1.3 mg, 0.0011%), 6 (112.8 mg, 0.0940%), 14 (56.3 mg, 0.0469%), and 15 (42.7 mg, 0.0356%). For compounds 1–4 a second preparative chromatography was carried out.
Citrinamine A (1): light-yellow powder; UV (DAD): λmax = 228, 270 nm; no CD effect (MeOH) was obtained (λ 210 to 300 nm); 1H and 13C NMR data in Table 1; HPLC/HR(+)ESIMS: tR = 16.4 min, m/z = 633.0330 [M + H]+ (calcd. for C22H23Br2N10O3, 633.0316), Δm = 2.9 ppm.
Citrinamine B (2): light-yellow powder; UV (DAD): λmax = 270 nm; no CD effect (MeOH) was obtained (λ 210 to 300 nm); 1H and 13C NMR data in Table 2; HPLC/HR(+)ESIMS: tR = 15.4 min, m/z = 740.0337 [M + H]+ (calcd. for C24H28Br2N11O5S, 740.0357), Δm = 2.7 ppm.
Citrinamine C (3): light-yellow powder; UV (DAD): λmax = 239, 272 nm; no CD effect (MeOH) was obtained (λ 210 to 300 nm); 1H and 13C NMR data in Supporting Information File 1, Table S1; HPLC/HR(+)ESIMS: tR = 17.7 min, m/z = 650.0493 [M + H]+ (calcd. for C23H26Br2N9O4, 650.0469), Δm = 2.9 ppm.
Citrinamine D (4): light-yellow powder; UV (DAD): λmax = 242, 269 nm; no CD effect (MeOH) was obtained (λ 210 to 300 nm); 1H and 13C NMR data in Supporting Information File 1, Table S1; HPLC/HR(+)ESIMS: tR = 16.7 min, m/z = 650.0470 [M + H]+ (calcd. for C23H26Br2N9O4, 650.0469), Δm = 0.2 ppm.
N-Methylagelongine (5): light-yellow powder; UV (DAD): λmax = 242, 274 nm; 1H and 13C NMR data in Table 3; HPLC/HR(+)ESIMS: tR = 11.9 min, m/z = 353.0135 [M + H]+ (calcd. for C14H14BrN2O4, 353.0131), Δm = 1.1 ppm.
Antimicrobial assay
As already described in [18] the antimicrobial activities were determined by agar diffusion tests using paper disks of 6 mm diameter soaked with 20 µL of the test compound in MeOH (1 mg/mL). The microorganisms were obtained from the HZI collection, grown on standard media, and cultured in liquid agar medium to a final OD of 0.01 (bacteria) or 0.1 (yeasts). Spores of fungi were collected from well-grown Petri dishes, which were rinsed with 10 mL of sterile H2O. One mL of the spore suspension was added to 100 mL of molten agar medium. Plates were incubated at 30 °C, and the diameters of resulting inhibition zones were measured after 1 and 2 days.
Cell proliferation assay
As already described in [18] L929 mouse fibroblasts were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and cultivated at 37 °C and 10% CO2 in DME medium (high glucose) supplemented with 10% fetal calf serum. Cell culture reagents were purchased from Life Technologies Inc. (GIBCO BRL). Growth inhibition was measured in microtiter plates. Aliquots of 120 µL of the suspended cells (50000/mL) were added to 60 µL of serial dilutions of the test compounds. After 5 days the growth was determined using an MTT assay [19].
Acknowledgements
Financial support from the Deutsche Forschungsgemeinschaft (DFG) (Ko 1314/5-1 and 5-2, DFG-Forschergruppe FOR 934) is gratefully acknowledged. Sponge collection was carried out by Dr. M. Assmann during a scientific expedition to the Bahamas in 2001. During this time the project was also sponsored by the DFG (Ko 1314/3-1 to 3-4). We would like to acknowledge the support of Prof. Dr. J. R. Pawlik (University of North Carolina, Wilmington, USA) who gave members of the Köck research group the opportunity to participate in scientific expeditions to the Bahamas (1998 to 2008). We further thank Dr. F. Sasse and B. Hinkelmann (Department of Chemical Biology, Helmholtz Centre for Infection Research, Braunschweig, Germany) for performing the bioassays.
References
-
Forte, B.; Malgesini, B.; Piutti, C.; Quartieri, F.; Scolaro, A.; Papeo, G. Mar. Drugs 2009, 7, 705–753. doi:10.3390/md7040705
Return to citation in text: [1] -
Kobayashi, J.; Ohizumi, Y.; Nakamura, H.; Hirata, Y. Experientia 1986, 42, 1176–1177. doi:10.1007/BF01941300
Return to citation in text: [1] -
Nakamura, H.; Ohizumi, Y.; Kobayashi, J.; Hirata, Y. Tetrahedron Lett. 1984, 25, 2475–2478. doi:10.1016/S0040-4039(01)81208-9
Return to citation in text: [1] -
Cafieri, F.; Fattorusso, E.; Mangoni, A.; Taglialatela-Scafati, O. Tetrahedron Lett. 1996, 37, 3587–3590. doi:10.1016/0040-4039(96)00629-6
Return to citation in text: [1] -
Uemoto, H.; Tsuda, M.; Kobayashi, J. J. Nat. Prod. 1999, 62, 1581–1583. doi:10.1021/np9902542
Return to citation in text: [1] -
Aiello, A.; D'Esposito, M.; Fattorusso, E.; Menna, M.; Müller, W. E. G.; Perović-Ottstadt, S.; Schröder, H. C. Bioorg. Med. Chem. 2006, 14, 17–24. doi:10.1016/j.bmc.2005.07.057
Return to citation in text: [1] -
Kobayashi, J.; Inaba, K.; Tsuda, M. Tetrahedron 1997, 53, 16679–16682. doi:10.1016/S0040-4020(97)10097-7
Return to citation in text: [1] -
Appenzeller, J.; Tilvi, S.; Martin, M.-T.; Gallard, J.-F.; El-bitar, H.; Dau, E. T. H.; Debitus, C.; Laurent, D.; Moriou, C.; Al-Mourabit, A. Org. Lett. 2009, 11, 4874–4877. doi:10.1021/ol901946h
Return to citation in text: [1] -
Kubota, T.; Araki, A.; Yasuda, T.; Tsuda, M.; Fromont, J.; Aoyama, K.; Mikami, Y.; Wälchli, M. R.; Kobayashi, J. Tetrahedron Lett. 2009, 50, 7268–7270. doi:10.1016/j.tetlet.2009.10.017
Return to citation in text: [1] -
Tilvi, S.; Moriou, C.; Martin, M.-T.; Gallard, J.-F.; Sorres, J.; Patel, K.; Petek, S.; Debitus, C.; Ermolenko, L.; Al-Mourabit, A. J. Nat. Prod. 2010, 73, 720–723. doi:10.1021/np900539j
Return to citation in text: [1] -
Cafieri, F.; Fattorusso, E.; Mangoni, A.; Taglialatela-Scafati, O.; Carnuccio, R. Bioorg. Med. Chem. Lett. 1995, 5, 799–804. doi:10.1016/0960-894X(95)00116-B
Return to citation in text: [1] [2] -
Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. J. Nat. Prod. 1996, 59, 501–503. doi:10.1021/np960113p
Return to citation in text: [1] -
Yasuda, T.; Araki, A.; Kubota, T.; Ito, J.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2009, 72, 488–491. doi:10.1021/np800645q
Return to citation in text: [1] [2] -
Olofson, A.; Yakushijin, K.; Horne, D. A. J. Org. Chem. 1997, 62, 7918–7919. doi:10.1021/jo9715682
Return to citation in text: [1] -
Endo, T.; Tsuda, M.; Okada, T.; Mitsuhashi, S.; Shima, H.; Kikuchi, K.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2004, 67, 1262–1267. doi:10.1021/np034077n
Return to citation in text: [1] [2] [3] -
Aiello, A.; D'Esposito, M.; Fattorusso, E.; Menna, M.; Müller, W. E. G.; Perović-Ottstadt, S.; Tsuruta, H.; Gulder, T. A. M.; Bringmann, G. Tetrahedron 2005, 61, 7266–7270. doi:10.1016/j.tet.2005.05.025
Return to citation in text: [1] [2] -
Grube, A.; Köck, M. J. Nat. Prod. 2006, 69, 1212–1214. doi:10.1021/np050408f
Return to citation in text: [1] -
Grube, A.; Assmann, M.; Lichte, E.; Sasse, F.; Pawlik, J. R.; Köck, M. J. Nat. Prod. 2007, 70, 504–509. doi:10.1021/np0603018
Return to citation in text: [1] [2] -
Mosmann, T. J. Immunol. Methods 1983, 65, 55–63. doi:10.1016/0022-1759(83)90303-4
Return to citation in text: [1]
| 16. | Aiello, A.; D'Esposito, M.; Fattorusso, E.; Menna, M.; Müller, W. E. G.; Perović-Ottstadt, S.; Tsuruta, H.; Gulder, T. A. M.; Bringmann, G. Tetrahedron 2005, 61, 7266–7270. doi:10.1016/j.tet.2005.05.025 |
| 11. | Cafieri, F.; Fattorusso, E.; Mangoni, A.; Taglialatela-Scafati, O.; Carnuccio, R. Bioorg. Med. Chem. Lett. 1995, 5, 799–804. doi:10.1016/0960-894X(95)00116-B |
| 16. | Aiello, A.; D'Esposito, M.; Fattorusso, E.; Menna, M.; Müller, W. E. G.; Perović-Ottstadt, S.; Tsuruta, H.; Gulder, T. A. M.; Bringmann, G. Tetrahedron 2005, 61, 7266–7270. doi:10.1016/j.tet.2005.05.025 |
| 1. | Forte, B.; Malgesini, B.; Piutti, C.; Quartieri, F.; Scolaro, A.; Papeo, G. Mar. Drugs 2009, 7, 705–753. doi:10.3390/md7040705 |
| 5. | Uemoto, H.; Tsuda, M.; Kobayashi, J. J. Nat. Prod. 1999, 62, 1581–1583. doi:10.1021/np9902542 |
| 15. | Endo, T.; Tsuda, M.; Okada, T.; Mitsuhashi, S.; Shima, H.; Kikuchi, K.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2004, 67, 1262–1267. doi:10.1021/np034077n |
| 4. | Cafieri, F.; Fattorusso, E.; Mangoni, A.; Taglialatela-Scafati, O. Tetrahedron Lett. 1996, 37, 3587–3590. doi:10.1016/0040-4039(96)00629-6 |
| 15. | Endo, T.; Tsuda, M.; Okada, T.; Mitsuhashi, S.; Shima, H.; Kikuchi, K.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2004, 67, 1262–1267. doi:10.1021/np034077n |
| 3. | Nakamura, H.; Ohizumi, Y.; Kobayashi, J.; Hirata, Y. Tetrahedron Lett. 1984, 25, 2475–2478. doi:10.1016/S0040-4039(01)81208-9 |
| 14. | Olofson, A.; Yakushijin, K.; Horne, D. A. J. Org. Chem. 1997, 62, 7918–7919. doi:10.1021/jo9715682 |
| 2. | Kobayashi, J.; Ohizumi, Y.; Nakamura, H.; Hirata, Y. Experientia 1986, 42, 1176–1177. doi:10.1007/BF01941300 |
| 15. | Endo, T.; Tsuda, M.; Okada, T.; Mitsuhashi, S.; Shima, H.; Kikuchi, K.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2004, 67, 1262–1267. doi:10.1021/np034077n |
| 11. | Cafieri, F.; Fattorusso, E.; Mangoni, A.; Taglialatela-Scafati, O.; Carnuccio, R. Bioorg. Med. Chem. Lett. 1995, 5, 799–804. doi:10.1016/0960-894X(95)00116-B |
| 13. | Yasuda, T.; Araki, A.; Kubota, T.; Ito, J.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2009, 72, 488–491. doi:10.1021/np800645q |
| 18. | Grube, A.; Assmann, M.; Lichte, E.; Sasse, F.; Pawlik, J. R.; Köck, M. J. Nat. Prod. 2007, 70, 504–509. doi:10.1021/np0603018 |
| 8. | Appenzeller, J.; Tilvi, S.; Martin, M.-T.; Gallard, J.-F.; El-bitar, H.; Dau, E. T. H.; Debitus, C.; Laurent, D.; Moriou, C.; Al-Mourabit, A. Org. Lett. 2009, 11, 4874–4877. doi:10.1021/ol901946h |
| 9. | Kubota, T.; Araki, A.; Yasuda, T.; Tsuda, M.; Fromont, J.; Aoyama, K.; Mikami, Y.; Wälchli, M. R.; Kobayashi, J. Tetrahedron Lett. 2009, 50, 7268–7270. doi:10.1016/j.tetlet.2009.10.017 |
| 10. | Tilvi, S.; Moriou, C.; Martin, M.-T.; Gallard, J.-F.; Sorres, J.; Patel, K.; Petek, S.; Debitus, C.; Ermolenko, L.; Al-Mourabit, A. J. Nat. Prod. 2010, 73, 720–723. doi:10.1021/np900539j |
| 13. | Yasuda, T.; Araki, A.; Kubota, T.; Ito, J.; Mikami, Y.; Fromont, J.; Kobayashi, J. J. Nat. Prod. 2009, 72, 488–491. doi:10.1021/np800645q |
| 19. | Mosmann, T. J. Immunol. Methods 1983, 65, 55–63. doi:10.1016/0022-1759(83)90303-4 |
| 7. | Kobayashi, J.; Inaba, K.; Tsuda, M. Tetrahedron 1997, 53, 16679–16682. doi:10.1016/S0040-4020(97)10097-7 |
| 6. | Aiello, A.; D'Esposito, M.; Fattorusso, E.; Menna, M.; Müller, W. E. G.; Perović-Ottstadt, S.; Schröder, H. C. Bioorg. Med. Chem. 2006, 14, 17–24. doi:10.1016/j.bmc.2005.07.057 |
| 12. | Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. J. Nat. Prod. 1996, 59, 501–503. doi:10.1021/np960113p |
| 18. | Grube, A.; Assmann, M.; Lichte, E.; Sasse, F.; Pawlik, J. R.; Köck, M. J. Nat. Prod. 2007, 70, 504–509. doi:10.1021/np0603018 |
© 2015 Cychon et al; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (http://www.beilstein-journals.org/bjoc)