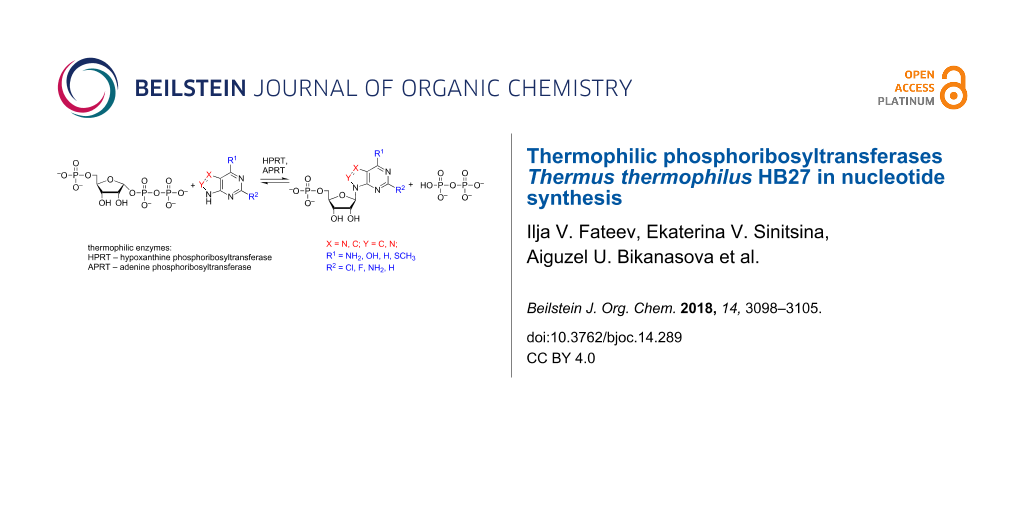
Abstract
Phosphoribosyltransferases are the tools that allow the synthesis of nucleotide analogues using multi-enzymatic cascades. The recombinant adenine phosphoribosyltransferase (TthAPRT) and hypoxanthine phosphoribosyltransferase (TthHPRT) from Thermus thermophilus HB27 were expressed in E.coli strains and purified by chromatographic methods with yields of 10–13 mg per liter of culture. The activity dependence of TthAPRT and TthHPRT on different factors was investigated along with the substrate specificity towards different heterocyclic bases. The kinetic parameters for TthHPRT with natural substrates were determined. Two nucleotides were synthesized: 9-(β-D-ribofuranosyl)-2-chloroadenine 5'-monophosphate (2-Сl-AMP) using TthAPRT and 1-(β-D-ribofuranosyl)pyrazolo[3,4-d]pyrimidine-4-one 5'-monophosphate (Allop-MP) using TthНPRT.
Graphical Abstract
Introduction
Bacterial phosphoribosyltransferases are used in multi-enzymatic cascades that perform nucleotide synthesis de novo [1,2]. Recently, we reported on the possibility of cascade synthesis, where enzymes of thermophilic microorganisms Thermus thermophilus HB27 (phosphoribosylpyrophosphate synthetase – PRPPS and adenine phosphoribosyltransferase – APRT) and Thermus sp. 2.9 (ribokinase – RK) carry out successive transformations of ribose and adenine heterocyclic bases into the corresponding nucleotides (Figure 1). The use of thermophilic phosphoribosyltransferases allows carrying out reactions at a higher temperature, so the concentrations of heterocyclic bases can be increased [1-3].
Figure 1: A multi-enzymatic synthesis of modified adenosine -5'-monophosphates.
Figure 1: A multi-enzymatic synthesis of modified adenosine -5'-monophosphates.
There is great interest in the development of multi-enzymatic cascades [4-9] for the preparation of nucleosides and nucleotides due to the regio- and stereospecificity of enzymes [4,10,11], performing metabolic transformations of substrates. Phosphoribosyltransferases are increasingly being widely used as key enzymes in multi-enzymatic systems [2]. The substrate specificity of APRT limits the number of possible nucleotides that can be synthesized. Thus, for Thermus thermophilus HB27 APRT (TthAPRT), nucleotide synthesis is limited to the closest structural homologs of adenine (Table 1) [1].
Table 1: Substrates for TthAPRT.
| Base | Conversion into nucleotide (24 h, %)a |
|---|---|
| 2,6-diaminopurine | 16.80 |
| 2-chloroadenine | 97.58 |
| 2-fluoroadenine | 36.50 |
| adenine | 50.02 |
| 2-methoxyadenine | 60.88 |
| N1-methyladenine | 78.2 |
| N6-benzyladenine | 1.87 |
| 2-aminobenzimidazole | 0.09 |
aReaction mixtures (0.5 mL, 20 mM Tris-HCl, pH 8.0, 75 °C) contained 0.4 mM heterocyclic base, 0.4 mM PRPP, 0–5 mM MgCl2, 1.25 μg TthAPRT [1].
Unfortunately, 1,2,4-triazole-3-carboxamide, its analogues, guanine, hypoxanthine, and 7-deazapurins are not substrates for TthAPRT. This severely limits the usability of multi-enzymatic cascades in the synthesis of nucleotides, including the modified ones.
To expand the possible repertoire of nucleotides that could be synthesized, we obtained the recombinant form of hypoxathine phosphoribosyltransferase Thermus thermophilus (TthHPRT), investigated its substrate specificity and optimal conditions for catalytic activity, and determined the kinetic parameters of the enzyme. A comparative study of the substrate specificity of TthAPRT and TthHPRT was performed to determine the usability of thermophilic transferases in nucleotide synthesis. A scheme of purine nucleotide synthesis using TthAPRT and TthHPRT is shown in Figure 2.
Figure 2: Nucleotide synthesis using phosphoribosyltransferases.
Figure 2: Nucleotide synthesis using phosphoribosyltransferases.
Results and Discussion
Genes TT_RS08985 and TT_RS06315 from T. thermophilus HB27, coding TthHPRT and TthAPRT, were cloned into expression plasmid vectors pET 23a+ and pET 23d+, respectively. The resulting recombinant plasmid pER-TthHPRT contained fusion gene HPRT-HisTag coding TthHPRT with a C-terminal His-Tag. The resulting recombinant plasmid pER-TthAPRT contained the gene APRT coding TthAPRT without any additional sequences. Nucleotide sequences of the cloned genes were verified by sequencing. The codone GGG→AGG substitution corresponding to amino-acid Arg27Gly replacement was found in the gene encoding the TthHPRT.
The screening of available producer strains was performed to find strains, which produce target enzymes in soluble form. The resulting strains E. coli BL21(DE3)/pER- TthAPRT and E. coli C3030/pER- TthHPRT produced enzymes mainly in soluble form (>80%).
The established procedure for isolation and purification of TthHPRT includes heat treatment, immobilized metal affinity chromatography, final size-exclusion chromatography, and concentration. For TthAPRT, the protocol include heat treatment, anion exchange chromatography, hydrophobic chromatography, final size-exclusion chromatography, and concentration. Yields of both transferases were no less than 10–13 mg per liter of culture, with a purity of about 96% (as determined by SDS-PAGE).
The influence of temperature and Mg2+ concentration on the activity of TthHPRT was investigated. The results were compared with data for adenine phosphoribosyltransferase Thermus thermophilus, obtained earlier [1].
The TthAPRT is active over a wide temperature range (Figure 3). A maximal activity of TthHPRT (1.1 unit/mg) is observed at 60 °C. The activity at 36 °C is 5% from the maximal one and at 90 °C it is 3% from the maximal one. It is interesting, that TthAPRT shows its maximal activity at 75 °C.
![[1860-5397-14-289-3]](/bjoc/content/figures/1860-5397-14-289-3.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 3: Dependence of TthHPRT and TthAPRT activity on temperature (reaction mixtures (0.5 mL) contained 20 mM Tris-HCl, pH 8.0, 1 mM 5-phosphoribosyl-1-α-pyrophosphate, and 5 mM MgCl2; in the case of TthHPRT mixtures contained 1 mM hypoxanthine and 0.18 μg of enzyme, in the case of TthAPRT – 1 mM adenine and 0.125 μg of enzyme).
Figure 3: Dependence of TthHPRT and TthAPRT activity on temperature (reaction mixtures (0.5 mL) contained 20 ...
The influence of the magnesium ion concentration on the TthHPRT activity is nonlinear. The activity increases rapidly while the magnesium chloride concentration increases from 0 to 1 mM (Figure 4). Further increasing of the concentration (up to 5 mM) does not increase the activity significantly. Since the reaction rate increases rapidly with increasing the magnesium chloride concentration to values equivalent to the concentration of 5-phosphoribosyl-α-1-pyrophosphate (1 mM), it can be assumed that the presence of magnesium ions promotes the proper spatial orientation of the substrate. The reaction also proceeds in the absence of magnesium ions in solution. A similar dependence is observed for TthAPRT.
![[1860-5397-14-289-4]](/bjoc/content/figures/1860-5397-14-289-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: Dependence of TthHPRT and TthAPRT activity on the Mg2+ concentration (reaction mixtures (0.5 mL) contained 20 mM Tris-HCl, pH 8.0, 1 mM 5-phosphoribosyl-1-α-pyrophosphate, and 0–5 mM MgCl2; in case of TthHPRT mixtures contained 1 mM hypoxanthine and 0.18 μg of enzyme, in case of TthAPRT – 1 mM adenine and 0.125 μg of enzyme; reactions were performed at 70 °C; 100% – activity at 1 mM concentration).
Figure 4: Dependence of TthHPRT and TthAPRT activity on the Mg2+ concentration (reaction mixtures (0.5 mL) co...
After optimization of the reaction conditions, kinetic parameters for TthHPRT were determined (Table 2).
Table 2: Kinetic parameters of inosine-5'-monophosphate and guanosine-5'-monophosphate synthesis using HPRT from various organisms.
| Substrate | Km, μМ | Vmax, μmol/min·mg | kcat, 1/s | kcat/Km, 1/M·s |
|---|---|---|---|---|
| Thermus thermophilus HB27 | ||||
| hypoxanthine | 13 ± 4 | 28 ± 9 | 9 ± 3 | 6.9 × 105 |
| guanine | 28 ± 9 | 6 ± 2 | 2.0 ± 0.7 | 7.1 × 104 |
| PRPP | 220 ± 60 | 17 ± 5 | 6 ± 2 | 2.7 × 104 |
| Thermus thermophilus HB8 [12] | ||||
| hypoxanthine | 3.9 ± 1.5 | – | 9.1 ± 0.8 | – |
| guanine | 7.4 ± 1.7 | – | 18 ± 1 | – |
| PRPP | 68 ± 18 | – | 20 ± 2 | – |
| Homo sapiens [13] | ||||
| hypoxanthine | 3.8 ± 0.3 | – | 2.6 ± 0.6 | 7 × 105 |
| PRPP | 19.1 ± 1.6 | – | 2.5 ± 0.05 | 1 × 105 |
| Escherichia coli [14] | ||||
| Hypoxanthine | 37 | – | – | – |
| PRPP | 330 | – | – | – |
Based on the Km values, the affinity of 5-phosphoribosyl-α-1-pyrophosphate for the active site is much lower than that of heterocyclic bases. The similar situation we observed for TthAPRT [1]. Comparison of the synthesis rates of inosine-5'-monophosphate and guanosine-5`-monophosphate showed that the first is synthesized 4.6 times faster. The literature data for similar enzymes (see Table 2) confirm a poor affinity of PRPP to the active site: Km for hypoxanthine is 17 fold less then for PRPP, although for the human enzyme Km is only 5 fold less. Comparing two enzymes from different strains of Thermus thermophilus, we can conclude that TthAPRT from HB8 (in contrast with HB27), synthesizes guanosine-5`-monophosphate faster. This may be due to the difference in reaction conditions. Kinetic data are displayed by double reciprocal plot (Figure 5). Determination of substrate specificity of TthHPRT was performed in comparative experiments with TthAPRT. The process of nucleotide synthesis was monitored by a liquid chromatography–mass spectrometry analysis of the reaction mixture.
![[1860-5397-14-289-5]](/bjoc/content/figures/1860-5397-14-289-5.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 5: Lineweaver–Burk plot for synthesis of inosine-5'-monophosphate and guanosine-5'-monophosphate.
Figure 5: Lineweaver–Burk plot for synthesis of inosine-5'-monophosphate and guanosine-5'-monophosphate.
The data is presented in the Table 3. As expected, TthHPRT is specific to 6-oxopurines, while TthAPRT is specific to 6-aminopurines. Both enzymes do not recognize thymine as a substrate. This is consistent with data that pyrimidine heterocyclic bases are substrates for uracyl phosphoribosyltransferase and orotate phosphoribosyltransferase only [2]. Unfortunately, we did not find any product in reactions with compounds based on 1,2,4-triazole-3-carboxamide, which was also observed for E. сoli HPRT [15,16]. However, allopurinol and 8-azaguanine are substrates for TthHPRT, and 2-chloroadenine is a substrate for TthAPRT. For 2-chloroadenine and 8-azaguanine, reaction at a higher temperature is preferable because of their low solubility in water (less than 1 mM at 37 °C). Interestingly, allopurinol proved to be a good substrate for both TthHPRT and TthAPRT, unlike hypoxanthine, which differs only in the position of one of the nitrogen atoms. Probably, the presence of nitrogen atom at C7 position of purine heterocycles plays an important role in reactions catalyzed by phosphoribosyltransferase, and also affects the substrate properties of TthHPRT and TthAPRT.
Table 3: Substrate specifity of TthHPRT and TthAPRT.
| Base | Conversion (24 h, %)a | |
|---|---|---|
| TthHPRT | TthAPRT | |
| adenine | 5.3 | 48.1 |
| hypoxanthine | 91.0 | 6.4 |
| guanine | 73.9 | 25.6 |
| 2-chloroadenine | 0 | 52.9 |
| 2-fluoroadenine | 0 | 31.1 |
| 6-mercapopurine | 85.1 | 4.8 |
| allopurinol | 39.3 | 57.4 |
| 8-azaguanine | 80.6 | 1.0 |
| thymine | 0 | 0 |
| 1,2,4-triazole-3-carboxamide | 0 | 0 |
| 1,2,4-triazole-3-carboxy-N-methylamide | 0 | 0 |
aReaction mixtures (0.125 mL, 20 mM Tris-HCl, pH 8.0, 60 °C) contained 0.5 mM heterocyclic base, 0.5 mM PRPP, 0.5 mM MgCl2, 5 mМ KH2PO4 and 0.4 μg of TthHPRT or TthAPRT.
Two nucleotides were synthesized using TthHPRT or TthAPRT (see Figure 6). Synthesis of 2-Cl-AMP was performed at 75 °C. This allowed to achieve a concentration of 0.5 mM of the initial 2-chloroadenine. The reaction progress was monitored by HPLC. After 2 days (the product content in the reaction mixture was 54%), the reaction mixture was concentrated and the desired product was isolated by column chromatography on ion-exchange sorbents (anion and then cation-exchange). The yield of 2-Сl-AMP was 37%. A second nucleotide (Allop-MP) was synthesized at a lower temperature (60 °C). After 2 days, the product content in the reaction mixture was 55%. The product was isolated in the same way, with a yield of 32%.
Figure 6: Synthesis of nucleotides 2Cl-AMP and Allop-MP using phosphoribosyltransferases TthAPRT or TthHPRT respectively.
Figure 6: Synthesis of nucleotides 2Cl-AMP and Allop-MP using phosphoribosyltransferases TthAPRT or TthHPRT r...
Conclusion
The recombinant adenine phosphoribosyltransferase and hypoxanthine phosphoribosyltransferase from Thermus thermophilus HB27 were purificated with yields no less than 10–13 mg per litre of culture. A comparative study of substrate specificity of these enzymes towards different heterocyclic bases was carried out and temperature-dependence and magnesium chloride concentration-dependence of enzymes activity were determined. TthHPRT can be used for the synthesis of nucleotides containing different purine derivatives including 8-aza- and 8-aza-7-deazapurine. The use of hypoxanthine and adenine transferases in multi-enzyme cascades significantly extends the spectrum of synthetic purine nucleotides. Two nucleotides were synthesized: 9-(β-D-ribofuranosyl)-2-chloroadenine 5'-monophosphate (2-Сl-AMP) using TthAPRT and 1-(β-D-ribofuranosyl)pyrazolo[3,4-d]pyrimidine-4-one 5'-monophosphate (Allop-MP) using TthНPRT with yields of 37% and 32%, respectively. Using of hypoxanthine and adenine transferases in multi-enzyme cascades significantly extends the spectrum of synthetic purine nucleotides.
Experimental
Tris-buffer, acetic acid, sodium chloride, glycerol, acrylamide, N,N'-bisacrylamide, ATP, bromophenol blue, agarose, EDTA, IPTG, ampicillin, sodium dodecylsulfate, imidazole and DMF were purchased from Panreac (Spain, Barselona). Ethanol was purchased from MedChemProm. Coomassie Brilliant Blue R-250 was purchased from Bio-Rad (USA, CA). Bacto yeast extract, bacto tryptone, and bacto agar were purchased from Becton Dickinson Biosciences (USA, NJ). NaOH and HCl were purchased from Merck (USA, MA). Sodium persulfate, TEMED, ethidium bromide, and sodium azide were purchased from Helicon (Russia). dNTP was purchased from Thermo Fisher Scientific (USA, MA). DTT, phenylmethylsulfonyl chloride, magnesium chloride, nickel sulfate, potassium dihydroorthophosphate, Ni-IDA sepharose, 5-phosphoribosyl-α-1-pyrophosphate and all bases (adenine, hypoxanthine, guanine, 2-chloroadenine, etc.) were purchased from Sigma-Aldrich (USA, MO).
Bacterial strains: a) E. coli C3030 [MiniF lysY (CamR) / fhuA2 lacZ::T7 gene1 [lon] ompT ahpC gal λatt::pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB sulA11 R(mcr-73::miniTn10--TetS)2 [dcm] R(zgb-210::Tn10 --TetS) endA1 Δgor ∆(mcrC-mrr)114::IS10] New England Biolabs (USA, MA), b) E.coli BL21(DE3) fhuA2 [lon] ompT gal (λ DE3) [dcm] ΔhsdS λ DE3 = λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 Δnin5.
Plasmid vector: pET 23a+, pET 23d+ (Merck Millipore, USA, MA).
Enzymes: NdeI, XhoI, NcoI, T4 DNA-ligase (Thermo Scientific, USA, MA), Encyclo-polymerase (Eurogen, Russia).
The protein concentration was determined by the Bradford method [17], using BSA as a standard.
Protein purity was determined by electrophoresis in a polyacrylamide gel under denaturing conditions [18].
Cloning and creation of producer strain: Genes TT_RS08985 and TT_RS06315, encoding TthHPRT and TthAPRT, respectively, were amplified on the genomic DNA template of the T. thermophilus HB27 strain by a polymerase chain reaction (PCR) using synthetic primers. The genes were cloned into the expression vectors pET-23a+ and pET-23d+ respectively. The E. coli strains BL21(DE3)/pER- TthAPRT and C3030/pER- TthHPRT produced the target enzymes mainly in soluble form (culturing conditions: 4 h grow at 37 °С after supplementing with 0.4 mM IPTG).
Isolation and purification of TthHPRT: A cell pellet was resuspended in 50 mM Tris-HCl, 200 mМ NaCl, and 1 mМ phenylmethylsulfonyl fluoride (PMSF) рН 8.0 (1:10 w/v). The cells were disrupted by sonication for 30 min at 20 kHz at +4 °С. The cell debris was pelleted by centrifugation at 12,000 rpm for 30 min at +4 °С. The cell lysate was heat-treated (65 °С, 10 min) and the pellet was removed by centrifugation. The supernatant was applied to a column XK 16/20 (GE Healthcare, USA) packed with Ni-IDA Sepharose (Sigma Aldrich, USA) pre-equilibrated with 50 mM Tris-HCl and 200 mM NaCl at pH 8.0. Ballast proteins were eluted with a solution, containing 50 mM Tris-HCl, 50 mM imidazole, and 200 mM NaCl, pH 8.0 (4 CV, flow rate 2 mL/min). The target enzyme was eluted with solution, contained 50 mM Tris-HCl, 250 mM imidazole, and 200 mM NaCl, pH 8.0 (4 CV, flow rate 2 mL/min). Pooled fractions were concentrated by a polysulfonic membrane PBGC 10 kDa (Millipore, USA). The resulting solution was applied to a column with HiLoad 16/60 Superdex 75pg (GE Healthcare, USA), equilibrated by 20 mM Tris-HCl, 50 mM NaCl, 0.04% NaN3, and 10% glycerol, pH 8.0. Fractions, containing the target enzyme with a purity of more than 96 %, were pooled and concentrated up to a concentration of 13 ± 1 mg/mL.
Isolation and purification of TthAPRT: Cell biomass disruption and heat-treatment was performed as described in section "Isolation and purification of TthHPRT". The resulting solution was diluted (2-fold) with solution, comtained 50 mM Tris-HCl, 2 M (NH4)2SO4, pH 8.0, and applied to column XK 16/20 packed with Phenyl Sepharose HP (GE Healthcare, USA). The column was eluted by linear gradient of (NH4)2SO4 (1.0 – 0 M, 12 CV, flow rate 2 mL/min). Fractions, contained the target enzyme, were pooled and concentrated on polysulphonic membrane PBGC 10 kDa. The resulting solution was applied to column with HiLoad 16/60 Superdex 200, equilibrated by 20 mM Tris-HCl, 50 mM NaCl, 0.04% NaN3, and 5% glycerol, pH 8.0. Fractions, contained the target enzyme with purity more than 96%, were pooled and concentrated up to concentration 12 ± 1 mg/mL.
Enzyme assay: Each reaction mixture (0.5 mL, 20 mM Tris-HCl, pH 8.0) contained 1 mM 5-phosphoribosyl-1-α-pyrophosphate, 1 mM hypoxanthine, 5 mM MgCl2, and hypoxanthine phosphoribosyltransferase Thermus thermophilus (0.18 μg). Reaction mixtures were incubated at 70 °C. Substrate and product quantities were determined using HPLC (Waters 1525, column Ascentis Express C18, 2.7 μm, 3.0 × 75 mm, eluent A 0.1% aqeous TFA, eluent B 0.1% TFA / 70% acetonitrile in water, detection at 254 nm, Waters 2489).
Kinetic parameters determination: Each reaction mixture (1.0 mL, 20 mM Tris-HCl, pH 8.0) contained 5 mM MgCl2, hypoxanthine phosphoribosyltransferase Thermus thermophilus (0.18 μg), and the following components: a) hypoxanthine (0.01–0.50 mM) or guanine (0.01–0.20 mM) and 1 mM 5-phosphoribosyl-1-α-pyrophosphate to determine Km and Vmax for hypoxanthine and guanine, and b) 5-phosphoribosyl-1-α-pyrophosphate (0.05–1.20 mM) and 0.50 mM hypoxanthine to determine Km and Vmax for 5-phosphoribosyl-1-α-pyrophosphate. Reaction mixtures were incubated at 70 °C for 2 min. Product quantities were determined as described in the "Enzyme assay" section. Each experiment was repeated three times. Kinetic parameters were determined by nonlinear regression analysis using SciDAVis v0.2.4 software (free software, web site: scidavis.sourceforge.net). Catalytic constants (kcat) were calculated per 1 subunit (20.3 kDa, calculated based on amino acid sequence).
Mass spectra were measured on an Agilent 6224, ESI-TOF, LC/MS (USA) in positive ion mode (ESI), LCQ Fleet ion trap mass spectrometer (Thermo Electron, USA) and Agilent 1100 LC/MSD VL (Agilent Technologies) equipped an APCI and ESI source (positive and negative mode of ionization), 1100 DAD and ELSD PL-ELS 1000 (Polymer Laboratories).
Nucleotides synthesis
9-(β-D-Ribofuranosyl)-2-chloroadenine 5'-monophosphate (2-Сl-AMP): 2-Chloroadenine (17 mg, 0.10 mmol) was dissolved in water (203 mL) under stirring and heating at 90 °C , and after cooling to 70 °C, magnesium chloride hexahydrate (41 mg, 0.21 mmol) and potassium dihydroorthophosphate (276 mg, 2.03 mmol) were added. The pH of the solution was adjusted to 8.0 by 2 N potassium hydroxide. The pentasodium salt of 5-phosphoribosyl-α-1-pyrophosphate (70 mg, 0.14 mmol) and TthAPRT (5 units) were added, and the reaction mixture was incubated at 75 °C for 2 days; the reaction progress was monitored by HPLC. The reaction mixture was neutralized with 2 N hydrochloric acid and concentrated in vacuo to ca. 10 mL. The precipitate was filtered off, the filtrate was applied to the column with DEAE-Toyopearl 650C, bicarbonate form, 40 × 140 mm, and the product was eluted with triethylammonium bicarbonate (0.1 M). Fractions were concentrated in vacuo to ca. 10 mL, applied to the column with CM-Sephadex C-25, sodium form, 20 × 160 mm, and the product was eluted with water to give, after evaporation and drying in vacuo under P2O5, 16 mg (0.037 mmol; 37%) of 9-(β-D-ribofuranosyl)-2-chloroadenine 5'-monophosphate of 99% purity (HPLC). HRMS (ESI+): m/z [M + H]+ calcd for C10H13N5O7P1Cl1: 382.0315; found, 382.0353; [2M + H]+, found, 763.0606; [Base + H]+, found, 170.0244; 1Н NMR (DMSО-d6) δ ppm) 8.52 (s, 1Н, H8), 7.78 (br. s., 2H, NH2), 5.83 (d, J1’,2’ = 6 Hz, 1Н, H1’), 4.61 (m, 1Н, H2’), 4.23 (m, 1Н, H3’), 4.06 (m, 1Н, H4’), 3.84 (m, 2Н, H5a’, H5b’) ppm; 13C NMR (DMSО-d6) δ 156.61 (C2 or C6), 153.01 (C6 or C2), 150.73 (C4), 139.68 (C8), 117.57 (C5), 86.40 (C1’), 84.52 (C4’), 74.13 (C2’), 71.06 (C3’), 3.94 (C5’) ppm; 15N NMR (DMSО-d6) δ 242.7 (N7), 171.3 (N9), 86.84 (NH2) ppm.
1-(β-D-Ribofuranosyl)pyrazolo[3,4-d]pyrimidine-4-one 5'-monophosphate (Allop-MP): Allopurinol (14 mg, 0.10 mmol) was dissolved in water (203 mL) under stirring and heating at 90 °C, and after cooling to 50 °C, magnesium chloride hexahydrate (41 mg, 0.21 mmol) and potassium dihydroorthophosphate (276 mg, 2.03 mmol) were added. The pH of the solution was adjusted to 8.0 with 2 N potassium hydroxide. The pentasodium salt of 5-phosphoribosyl-α-1-pyrophosphate (70 mg, 0.14 mmol) and TthHPRT (5 units) were added, and the reaction mixture was incubated at 60 °C for 2 days; the reaction progress was monitored by HPLC. The reaction mixture was neutralized by 2 N hydrochloric acid and concentrated in vacuo to ca. 10 mL. The precipitate was filtered off, the filtrate was placed on the column with DEAE-Toyopearl 650C, bicarbonate form, 40 × 140 mm, and the product was eluted with triethylammonium bicarbonate (0.2 M). Fractions were concentrated in vacuo to ca. 10 mL, applied to the column with CM-Sephadex C-25, sodium form, 20 × 160 mm, and the product was eluted with water to give, after evaporation and drying in vacuo under P2O5, 11 mg (0.032 mmol; 32%) of 1-(β-D-ribofuranosyl)pyrazolo[3,4-d]pyrimidine-4-one 5'-monophosphate of 97% purity (HPLC). HRMS (ESI+): m/z [M + H]+ calcd for C10H13N4O8P1: 349.0545; found, 349.0520; [2M + H]+, found, 697.0952; [3M + H]+, found, 1045.1374; [Base + H]+ found, 137.0453; 1Н NMR (DMSО-d6) δ 12.44 (br. s, 1H, NH), 8.15 (s, 1H, H3), 8.13 (s, 1H, H6), 6.06 (d, J = 4.1 Hz, 1H, H1’), 4.56 (dt, 1H, H2’, J = 4.54; <0.5), 4.31 (t, 1H, H3’, J = 4.8), 4.04 (m, 1H, H4’), 3.85 (ddd, J = 11.0, 7.6; 6.2 Hz, 1H, H5’a), 3.66 (ddd, J = 11.0, 7.2, 6.1 Hz 1H, H5’b) ppm; 13C NMR (DMSО-d6) δ 157.03 (C4), 152.90 (C7a), 148.53 (C6), 135.38 (C3), 106.06 (C4a), 88.16 (C1’), 83.27 (C4’), 73.39 (C2’), 71.38 (C3’), 64.76 (C5’) ppm; 15N NMR (DMSО-d6) δ 302.8 (N2), 210.6 (N7), 204.9 (N1), 171.1 (N5).
Abbreviations
APRT – adenine phosphoribosyltransferase; HPRT – hypoxathine phosphoribosyltransferase; PRPPS – phosphoribosylpyrophosphate synthetase; RK – ribokinase; Tth - Thermus thermophilus; 2-Сl-AMP – 9-(β-D-ribofuranosyl)-2-chloroadenine 5'-monophosphate; Allop-MP – 1-(β-D-ribofuranosyl)-pyrazolo[3,4-d]pyrimidine-4-one 5'-monophosphate
Supporting Information
| Supporting Information File 1: Detailed analysis of mass spectrometry and NMR data. | ||
| Format: PDF | Size: 1.2 MB | Download |
References
-
Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90.
Return to citation in text: [1] [2] [3] [4] [5] [6] -
Del Arco, J.; Fernández-Lucas, J. Curr. Pharm. Des. 2017, 23, 1–15. doi:10.2174/1381612823666171017165707
Return to citation in text: [1] [2] [3] [4] -
Taran, S. A.; Verevkina, K. N.; Feofanov, S. A.; Miroshnikov, A. I. Russ. J. Bioorg. Chem. 2009, 35, 739–745. doi:10.1134/s1068162009060107
Return to citation in text: [1] -
Mikhailopulo, I. A. Curr. Org. Chem. 2007, 11, 317–335. doi:10.2174/138527207780059330
Return to citation in text: [1] [2] -
Mikhailopulo, I. A.; Miroshnikov, A. I. Mendeleev Commun. 2011, 21, 57–68. doi:10.1016/j.mencom.2011.03.001
Return to citation in text: [1] -
Wintersberger, E. Biochem. Soc. Trans. 1997, 25, 303–308. doi:10.1042/bst0250303
Return to citation in text: [1] -
Tesmer, J. J.; Klem, T. J.; Deras, M. L.; Davisson, V. J.; Smith, J. L. Nat. Struct. Biol. 1996, 3, 74–86. doi:10.1038/nsb0196-74
Return to citation in text: [1] -
Kim, M.-J.; Whitesides, G. M. Appl. Biochem. Biotechnol. 1987, 16, 95–108. doi:10.1007/bf02798359
Return to citation in text: [1] -
Scism, R. A.; Stec, D. F.; Bachmann, B. O. Org. Lett. 2007, 9, 4179–4182. doi:10.1021/ol7016802
Return to citation in text: [1] -
Mikhailopulo, I. A.; Miroshnikov, A. I. ActaNaturae 2010, 2, 36–59.
Return to citation in text: [1] -
Iglesias, L. E.; Lewkowicz, E. S.; Medici, R.; Bianchi, P.; Iribarren, A. M. Biotechnol. Adv. 2015, 33, 412–434. doi:10.1016/j.biotechadv.2015.03.009
Return to citation in text: [1] -
Kanagawa, M.; Baba, S.; Ebihara, A.; Shinkai, A.; Hirotsu, K.; Mega, R.; Kim, K.; Kuramitsu, S.; Sampei, G.-i.; Kawai, G. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2010, 66, 893–898. doi:10.1107/s1744309110023079
Return to citation in text: [1] -
Canyuk, B.; E-Wan, A.; Keawwijit, W.; Nualnoi, T.; Sirisatean, L.; Tansakul, P.; Tanthana, C. Nucleosides, Nucleotides Nucleic Acids 2008, 27, 894–899. doi:10.1080/15257770802146593
Return to citation in text: [1] -
Miller, R. L.; Ramsey, G. A.; Krenitsky, T. A.; Elion, G. B. Biochemistry 1972, 11, 4723–4731. doi:10.1021/bi00775a014
Return to citation in text: [1] -
Streeter, D. G.; Miller, J. P.; Robins, R. K.; Simon, L. N. Ann. N. Y. Acad. Sci. 1977, 284, 201–210. doi:10.1111/j.1749-6632.1977.tb21952.x
Return to citation in text: [1] -
Naesens, L.; Guddat, L. W.; Keough, D. T.; van Kuilenburg, A. B. P.; Meijer, J.; Voorde, J. V.; Balzarini, J. Mol. Pharmacol. 2013, 84, 615–629. doi:10.1124/mol.113.087247
Return to citation in text: [1] -
Laemmli, U. K. Nature 1970, 227, 680–685. doi:10.1038/227680a0
Return to citation in text: [1] -
Bradford, M. M. Anal. Biochem. 1976, 72, 248–254. doi:10.1016/0003-2697(76)90527-3
Return to citation in text: [1]
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 2. | Del Arco, J.; Fernández-Lucas, J. Curr. Pharm. Des. 2017, 23, 1–15. doi:10.2174/1381612823666171017165707 |
| 2. | Del Arco, J.; Fernández-Lucas, J. Curr. Pharm. Des. 2017, 23, 1–15. doi:10.2174/1381612823666171017165707 |
| 4. | Mikhailopulo, I. A. Curr. Org. Chem. 2007, 11, 317–335. doi:10.2174/138527207780059330 |
| 10. | Mikhailopulo, I. A.; Miroshnikov, A. I. ActaNaturae 2010, 2, 36–59. |
| 11. | Iglesias, L. E.; Lewkowicz, E. S.; Medici, R.; Bianchi, P.; Iribarren, A. M. Biotechnol. Adv. 2015, 33, 412–434. doi:10.1016/j.biotechadv.2015.03.009 |
| 18. | Bradford, M. M. Anal. Biochem. 1976, 72, 248–254. doi:10.1016/0003-2697(76)90527-3 |
| 4. | Mikhailopulo, I. A. Curr. Org. Chem. 2007, 11, 317–335. doi:10.2174/138527207780059330 |
| 5. | Mikhailopulo, I. A.; Miroshnikov, A. I. Mendeleev Commun. 2011, 21, 57–68. doi:10.1016/j.mencom.2011.03.001 |
| 6. | Wintersberger, E. Biochem. Soc. Trans. 1997, 25, 303–308. doi:10.1042/bst0250303 |
| 7. | Tesmer, J. J.; Klem, T. J.; Deras, M. L.; Davisson, V. J.; Smith, J. L. Nat. Struct. Biol. 1996, 3, 74–86. doi:10.1038/nsb0196-74 |
| 8. | Kim, M.-J.; Whitesides, G. M. Appl. Biochem. Biotechnol. 1987, 16, 95–108. doi:10.1007/bf02798359 |
| 9. | Scism, R. A.; Stec, D. F.; Bachmann, B. O. Org. Lett. 2007, 9, 4179–4182. doi:10.1021/ol7016802 |
| 2. | Del Arco, J.; Fernández-Lucas, J. Curr. Pharm. Des. 2017, 23, 1–15. doi:10.2174/1381612823666171017165707 |
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 2. | Del Arco, J.; Fernández-Lucas, J. Curr. Pharm. Des. 2017, 23, 1–15. doi:10.2174/1381612823666171017165707 |
| 3. | Taran, S. A.; Verevkina, K. N.; Feofanov, S. A.; Miroshnikov, A. I. Russ. J. Bioorg. Chem. 2009, 35, 739–745. doi:10.1134/s1068162009060107 |
| 15. | Streeter, D. G.; Miller, J. P.; Robins, R. K.; Simon, L. N. Ann. N. Y. Acad. Sci. 1977, 284, 201–210. doi:10.1111/j.1749-6632.1977.tb21952.x |
| 16. | Naesens, L.; Guddat, L. W.; Keough, D. T.; van Kuilenburg, A. B. P.; Meijer, J.; Voorde, J. V.; Balzarini, J. Mol. Pharmacol. 2013, 84, 615–629. doi:10.1124/mol.113.087247 |
| 12. | Kanagawa, M.; Baba, S.; Ebihara, A.; Shinkai, A.; Hirotsu, K.; Mega, R.; Kim, K.; Kuramitsu, S.; Sampei, G.-i.; Kawai, G. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2010, 66, 893–898. doi:10.1107/s1744309110023079 |
| 14. | Miller, R. L.; Ramsey, G. A.; Krenitsky, T. A.; Elion, G. B. Biochemistry 1972, 11, 4723–4731. doi:10.1021/bi00775a014 |
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 1. | Esipov, R. S.; Abramchik, Yu. A.; Fateev, I. V.; Konstantinova, I. D.; Kostromina, M. A.; Muravyova, T. I.; Artemova, K. G.; Miroshnikov, A. I. ActaNaturae 2016, 8, 82–90. |
| 13. | Canyuk, B.; E-Wan, A.; Keawwijit, W.; Nualnoi, T.; Sirisatean, L.; Tansakul, P.; Tanthana, C. Nucleosides, Nucleotides Nucleic Acids 2008, 27, 894–899. doi:10.1080/15257770802146593 |
© 2018 Fateev et al.; licensee Beilstein-Institut.
This is an Open Access article under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0). Please note that the reuse, redistribution and reproduction in particular requires that the authors and source are credited.
The license is subject to the Beilstein Journal of Organic Chemistry terms and conditions: (https://www.beilstein-journals.org/bjoc)