Search results
Search for "Python" in Full Text gives 25 result(s) in Beilstein Journal of Organic Chemistry.
Understanding the origin of stereoselectivity in the photochemical denitrogenation of 2,3-diazabicyclo[2.2.1]heptene and its derivatives with non-adiabatic molecular dynamics
Beilstein J. Org. Chem. 2025, 21, 2007–2020, doi:10.3762/bjoc.21.156

- enumerate all possible mechanistic pathways. Non-adiabatic molecular dynamics simulations We performed an NAMD simulation using the Fewest Switches Surface Hopping (FSSH) algorithm [92][93][94], with OpenMolcas [95][96] and with Python Rapid Artificial Intelligence Ab Initio Molecular Dynamics (Pyrai2MD
- mechanical calculations are performed in OpenMolcas using SA(4)-CASSCF(8,9)/ANO-S-VDZP. With the Python Rapid Artificial Intelligence Ab Initio Molecular Dynamics (PyRAI2MD) [97][98][99], we used FSSH to run the NAMD simulations. Trajectories were initiated from the S1 Franck–Condon (FC) region, with a total
Measuring the stereogenic remoteness in non-central chirality: a stereocontrol connectivity index for asymmetric reactions
Beilstein J. Org. Chem. 2025, 21, 1995–2006, doi:10.3762/bjoc.21.155
- generation of indices following the 3-step procedure using a prototypic program coded using Python or through coaching GPT-4.1. Stereoselective reactions were input as SMILES (for Python) or InChI (for GPT-4.1) of the starting materials and products, and the corresponding indices were generated automatically
- chirality derived from C=C bonds. Examples of reactions that establish planar chirality. Examples of reactions that establish “inherent” chirality. Parameterization of asymmetric reactions that establish axial chirality. Supporting Information Supporting Information File 24: The set of code (Python) and
Enhancing chemical synthesis planning: automated quantum mechanics-based regioselectivity prediction for C–H activation with directing groups
Beilstein J. Org. Chem. 2025, 21, 1171–1182, doi:10.3762/bjoc.21.94

- influence of DGs, we refer the reader to previous work done by Kromann et al. [15] and Ree and colleagues [16][17]. The there developed RegioSQM predicts the regioselectivity of reactions following the electrophilic aromatic substitution mechanism within seconds to minutes using a web interface or a Python
- interface, a web-based user interface, an API, and a stand-alone Python module for integration into more complex systems. For example, this workflow can be used in further molecular discovery and optimization to design specific directing groups that can facilitate the functionalization of remote C–H bonds
Emerging trends in the optimization of organic synthesis through high-throughput tools and machine learning
Beilstein J. Org. Chem. 2025, 21, 10–38, doi:10.3762/bjoc.21.3

- repository to be employed freely for research purposes. In addition to HappyTools, there are other available open-source Python packages to analyze chromatographic and spectroscopic data. A cross-platform Python package named Aston can be used to process both UV–vis and MS data. The open-source library is
- written using Python, NumPy, and SciPy and is openly hosted in an online repository [68]. Similarly, for processing chromatographic data from GC–FID, HPLC–UV, or HPLC–FD, packages are also available open source. Embedding these codes into HTE and ML workflow dramatically improves the efficiency and speed
- of the optimization processes. Liu et al. [69] developed a custom-built Python script to study the kinetics of carbonyldiimidazole-mediated amide formation by analyzing data from online HPLC and in-line FTIR-spectroscopic measurements. Their algorithm was able to automatically detect peaks from
Computational design for enantioselective CO2 capture: asymmetric frustrated Lewis pairs in epoxide transformations
Beilstein J. Org. Chem. 2024, 20, 2668–2681, doi:10.3762/bjoc.20.224

- fluoride ion, respectively. Volcanic 1.3.3, a Python package for the NaviCat platform, was used to generate 3D volcano plots, facilitating the identification of the most appropriate catalyst for the coupling reaction being considered [27]. Volcano plots Volcano plots are a visualisation of the Sabatier
Catalysing (organo-)catalysis: Trends in the application of machine learning to enantioselective organocatalysis
Beilstein J. Org. Chem. 2024, 20, 2280–2304, doi:10.3762/bjoc.20.196

Computational toolbox for the analysis of protein–glycan interactions
Beilstein J. Org. Chem. 2024, 20, 2084–2107, doi:10.3762/bjoc.20.180

- python framework that allows users to prepare carbohydrate structures for atomistic simulations of complex glycoproteins, glycolipids and carbohydrate polymers in the GROMACS force field format (see below). Polysaccharides can be prepared by using the prepreader.py tool, glycoproteins and glycolipids by
- registered to the web site. 4. Azahar [53]: freely available python-based plugin that permits to visualise, analyse and model glycans and glycoconjugates (https://pymolwiki.org/index.php/Azahar). 5. 3D-SNFG: It is a script integrated in the visual molecular dynamics (VMD) program [54] (see below) that allows
A fiber-optic spectroscopic setup for isomerization quantum yield determination
Beilstein J. Org. Chem. 2024, 20, 1684–1692, doi:10.3762/bjoc.20.150

- . The background power was therefore monitored to get an accurate power reading before turning on the LED. The raw data was then fitted with a simple polynomial baseline using a Python script (see Supporting Information File 1). After baseline subtraction, the average power and standard deviation for
- different python scripts (provided in Supporting Information File 1). The rate equations are ordinary differential equations (ODEs) and can thus be solved numerically using existing methods in the SciPy package [50]. An absorbance function was constructed, which takes the solution of the ODEs to model the
Monitoring carbohydrate 3D structure quality with the Privateer database
Beilstein J. Org. Chem. 2024, 20, 931–939, doi:10.3762/bjoc.20.83

- entry in the PDB [24] or in PDB-REDO [21]. For each structure in the PDB, the carbohydrate-containing chains are first identified before being validated using the suite of validation tools available within Privateer. Using the Python bindings available within the latest versions of Privateer, a
Optimizing reaction conditions for the light-driven hydrogen evolution in a loop photoreactor
Beilstein J. Org. Chem. 2024, 20, 74–91, doi:10.3762/bjoc.20.9
- shot under red light, which matched the absorption properties of the dye, to obtain optimal optical visualization. Digital images extracted from videos (see Supporting Information File 2) were processed with the Python OpenCV package to quantify the mixing time (see Supporting Information File 3
- , Python code) by following the color evolution in the videos from red towards black under the red-light environment [49]. ROI used as the working zone were defined on the images when these were processed to quantify the mixing time. The three rectangles shown in Figure 3 on frame at 1.2 s showed the
- time quantification. For each frame of the video, the average red channel value of all the pixels in the marked ROIs were calculated. The mixing time calculation is explained in Supporting Information File 1 and the Python code for the corresponding calculation is shown in Supporting Information File 3
Cytochrome P450 monooxygenase-mediated tailoring of triterpenoids and steroids in plants
Beilstein J. Org. Chem. 2022, 18, 1289–1310, doi:10.3762/bjoc.18.135
- on triterpenoids and steroids. Amino acid sequences (149 triterpenoid CYPs, 266 non-triterpenoid CYPs) were aligned using MUSCLE [92], and a neighbour-joining consensus tree of 1,000 bootstrap replicates was generated using the Jukes–Cantor model. The final tree was visualised in Python using the
Heterogeneous metallaphotoredox catalysis in a continuous-flow packed-bed reactor
Beilstein J. Org. Chem. 2022, 18, 1123–1130, doi:10.3762/bjoc.18.115
- time by automatically acquiring, processing and integrating the 19F NMR spectrum of the reaction mixture flowing in the spectrometer (see relevant code in Supporting Information File 3). In particular, the python packages flowchem [35] and nmrglue [36] were used to control the spectrometer and process
In-depth characterization of self-healing polymers based on π–π interactions
Beilstein J. Org. Chem. 2021, 17, 2496–2504, doi:10.3762/bjoc.17.166
- along the scratch (every 20 µm) were performed with a 10 µm Rockwell indenter with the following parameters: 3 mN normal force, 200 µm/min scratch speed, length 1600 µm (P1) or 600 µm (P2). For the visualization and evaluation of the scratch profile data recorded by the MST3, a Python-based GUI
Simulating the enzymes of ganglioside biosynthesis with Glycologue
Beilstein J. Org. Chem. 2021, 17, 739–748, doi:10.3762/bjoc.17.64
- obesity and insulin resistance [36]. Glycologue web application The Glycologue ganglioside simulator is available at https://glycologue.org/g/, along with the source code of the simulator in the Python programming language. Glycologue exports networks as SBML, for import into Copasi [37], CellDesigner [38
Semiautomated glycoproteomics data analysis workflow for maximized glycopeptide identification and reliable quantification
Beilstein J. Org. Chem. 2020, 16, 3038–3051, doi:10.3762/bjoc.16.253

- quantification in LaCyTools [16]. A python script was developed to facilitate this step (Supporting Information File 3). LaCyTools was chosen because it is open-source, can be applied for a large number of samples (thousands of samples in one study have been reported [19]), and allows data curation and
- data handling, making it an excellent tool for, e.g., clinical cohort analysis. In the current work, a python script was developed to streamline the connection between GlycopeptideGraphMS identification and LaCyTools quantification (Supporting Information File 3). All tools provided absolute values for
- quantification in LaCyTools (v 1.0.1) [16], the raw data were converted to the mzXML format by MSConvert. The generation of the LaCyTools analyte list was supported by an in-house Python (v 3.7.6) script (Supporting Information File 3), which converted a representative GlycopeptideGraphMS output to the required
Computational tools for drawing, building and displaying carbohydrates: a visual guide
Beilstein J. Org. Chem. 2020, 16, 2448–2468, doi:10.3762/bjoc.16.199

- for preparing carbohydrate structures for atomistic simulations of glycoproteins, carbohydrate polymers and glycolipids using GROMACS [70][71] In the form of Python scripts; the tools are used to prepare the system, which generally includes the processing of a.pdb file using the pdb2gmx tool
- computer code mostly written in C++ and Python languages. Rosetta is available to academic and commercial researchers through a license available at https://www.rosettacommons.org/software/license-and-download. The licence is free for academic users. The tool runs best on Linux or macOS platforms only. It
GlypNirO: An automated workflow for quantitative N- and O-linked glycoproteomic data analysis
Beilstein J. Org. Chem. 2020, 16, 2127–2135, doi:10.3762/bjoc.16.180

- -glycosylation; O-glycosylation; Python; Introduction Glycosylation is a key post-translational modification critical for protein folding and function in eukaryotes [1][2][3]. Diverse types of glycosylation are known, all involving modification of specific amino acid residues with complex carbohydrate
- robustness and throughput. Our workflow uses a collection of scripts built on an in-house sequence string handling library and the scientific Python data handling package pandas [24], and integrates outputs of two commonly used software packages in glycoproteomic MS data analysis, Proteome Discoverer (Thermo
- Python 3.8.3 with backward compatibility tested up to Python 3.6. Each Byonic output file was first iteratively prepared for linking with AUC information from the Proteome Discoverer output. Using a regular expression pattern provided by UniProtKB, the UniProtKB accession ID of each protein from the
Clustering and curation of electropherograms: an efficient method for analyzing large cohorts of capillary electrophoresis glycomic profiles for bioprocessing operations
Beilstein J. Org. Chem. 2020, 16, 2087–2099, doi:10.3762/bjoc.16.176

- the calibration algorithm see [16]. The calibrated electropherograms were then clustered. The clustering algorithm was implemented in-house using the SciPy python package. The clustering step consists of hierarchical clustering using a single linkage algorithm and forms flat clusters using the
In silico rationalisation of selectivity and reactivity in Pd-catalysed C–H activation reactions
Beilstein J. Org. Chem. 2020, 16, 1465–1475, doi:10.3762/bjoc.16.122
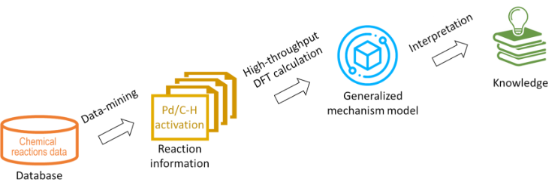
- modern supercomputer clusters [19]. The structures were generated by the Python module developed in house and explained in detail elsewhere [20]. Electronic energies were evaluated using Becke’s three-parameter hybrid B3LYP functional, while the molecular orbitals are expanded in triple-zeta all electron
- predictions. Employing the Python module [20] and OpenBabel executables [28], the 3D structures of the most stable conformers were generated from the 2D structure of a substrate. Subsequently, structures of all possible palladium intermediates representing both mechanisms (PA and SEAr) were built for each
Nanangenines: drimane sesquiterpenoids as the dominant metabolite cohort of a novel Australian fungus, Aspergillus nanangensis
Beilstein J. Org. Chem. 2019, 15, 2631–2643, doi:10.3762/bjoc.15.256

- A. calidoustus SF006504 (GCA_001511075.1), A. ochraceus fc-1 (GCA_004849945.1), A. parasiticus SU-1 (GCA_000956085.1), A. sclerotiorum HBR18 (GCA_000530345.1) and A. ustus 3.3904 (GCA_000812125.1). Homologous gene clusters were identified in these genomes using a custom Python script, named
- was visualised using another custom Python script named crosslinker (https://github.com/gamcil/crosslinker). Further details regarding clusterblaster and crosslinker are given in Supporting Information File 1. Structures of nanangenines 1–10 isolated from A. nanangensis. Putative nanangenine
A permutation approach to the assignment of the configuration to diastereomeric tetrads by comparison of experimental and ab initio calculated differences in NMR data
Beilstein J. Org. Chem. 2017, 13, 2478–2485, doi:10.3762/bjoc.13.245

- diastereomers of compounds 1–7. Supporting Information Supporting Information File 460: Plots of NMR spectra for new compounds, HSQC experiments for tetrad 1, supporting tables, complete reference [16], synthetic references for compounds 1–4, presentation of manual workflow and Python code for automated
The digital code driven autonomous synthesis of ibuprofen automated in a 3D-printer-based robot
Beilstein J. Org. Chem. 2016, 12, 2776–2783, doi:10.3762/bjoc.12.276
- one-pot three-step approach. The synthesis of ibuprofen could be achieved on different scales simply by adjusting the parameters in the robot control software. The software for controlling the synthesis robot was written in the python programming language and hard-coded for the synthesis of ibuprofen
- open source allowing the printer to be easily interfaced with user-developed modifications, allowing us to produce our own software for coordinating the 3D printing, liquid handling and reaction timing (see Scheme 2). The software to control our robotic platform was written in Python and the full
Mechanical stability of bivalent transition metal complexes analyzed by single-molecule force spectroscopy
Beilstein J. Org. Chem. 2015, 11, 817–827, doi:10.3762/bjoc.11.91

- constant velocities between 100 nm/s and 10 µm/s using a grid of different spots on the surface. Force–distance curves were processed as described in [27]. In short, signals were fitted according to the wormlike-chain model using Hooke, a Python-based force spectroscopy data analysis program [56]. Most
Integration of enabling methods for the automated flow preparation of piperazine-2-carboxamide
Beilstein J. Org. Chem. 2014, 10, 641–652, doi:10.3762/bjoc.10.56

- experience using the Python programming language [6] to control laboratory devices [5]. This language claims to be ideal for rapid development and the use of free software fosters collaboration [7], enabling technology to be transferred without the large initial set-up costs typically involved with
- and monitoring system being developed within our group [26], which can carry out a programmed sequence of operations written using the Python™ language. There is also a remote interface for observing the status of an ongoing reaction in real-time. In common with the industrial use of process
Camera-enabled techniques for organic synthesis
Beilstein J. Org. Chem. 2013, 9, 1051–1072, doi:10.3762/bjoc.9.118
- environment. However, the availability of powerful open-source image processing software such as the Python Imaging Library [68] (PIL, an image processing library for the Python [69] programming language) and OpenCV [70] (a C++ library for creating real-time computer vision applications with bindings for the
- Python language [71]) enables powerful image-recognition logic to be harnessed for a low cost and by nonspecialists. Access to these libraries is only one contributing factor: such software projects are often well documented with a number of examples, and the internet provides a medium for rapidly
- sharing and recycling code and applications. Another important aspect of software libraries such as these is that whilst they use highly efficient C or C++ code to perform processor-intensive calculations, the “bindings” allow their full functionality to be harnessed by using languages such as Python




























































































































































































































































